Regulation of chromatin stability in mammalian cells
Transcription and replication constantly disturb the contact of histones with DNA and may "degrade" chromatin. However, chromatin is not "degraded" even in tumor cells, in which both processes are significantly accelerated.
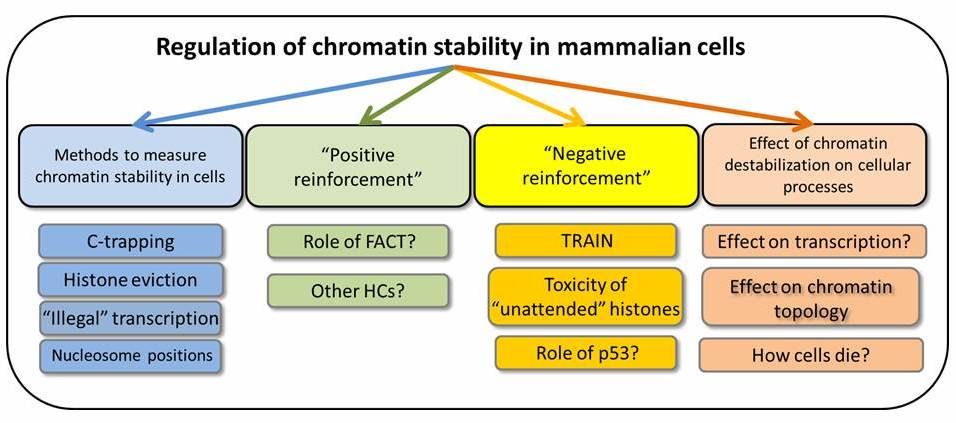
This suggests that there should be a system to monitor and maintain chromatin composition to avoid loss of epigenetic marks and differentiation. We propose that this may be achieved via two ways:
- Positive reinforcement or constant repair of the chromatin; and
- Negative reinforcement or elimination of cells with damaged chromatin.
Although neither of these systems is clearly described, our data suggests evidence of both.
We showed that FACT is one of the major sensors of chromatin destabilization. It detects nucleosome disassembly and binds destabilized nucleosomes via SPT16 subunits, and DNA undergoing super-helical transitions due to nucleosome loss via SSRP1 subunits.
There are at least two DNA binding domains on c-terminus of SSRP1, HMG box which can bind bent or cruciform DNA and CID which binds left-handed Z-DNA.
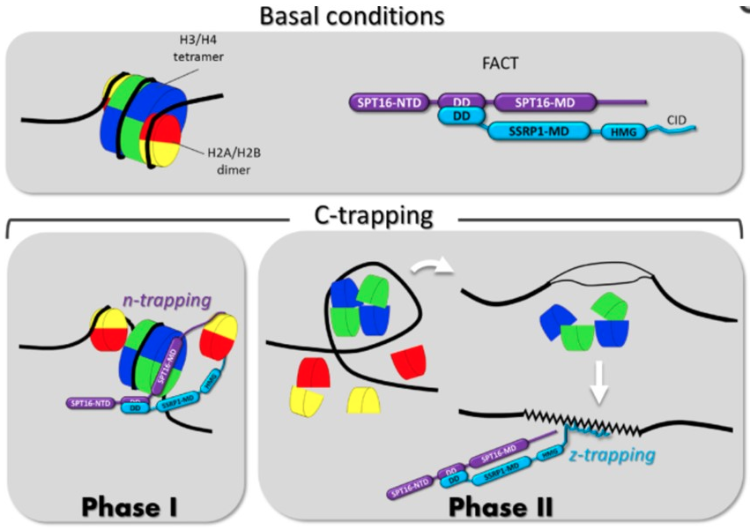
How FACT detects chromatin destabilization: model of the mechanism of c-trapping
In the image below, the upper panel shows the scheme of the nucleosome with a standard color code for core histones (H2A-yellow, H2B – red, H3 – blue, H4 – green) and the domain structure of FACT subunits (NTD – N-terminal domain, DD – dimerization domain, MD – middle domain).
The lower panels show two phases of c-trapping:
- n(nucleosome)-trapping that occurs via the SPT16 subunit binding to the hexasome of the partially uncoiled nucleosome when one molecule of CBL0137 is bound per ∼10–100 bp of DNA; and
- z(Z-DNA)-trapping that occurs via SSRP1 subunit binding to DNA when the nucleosome is disassembled upon binding of one or more molecules of CBL0137 to every 10 bp of DNA.
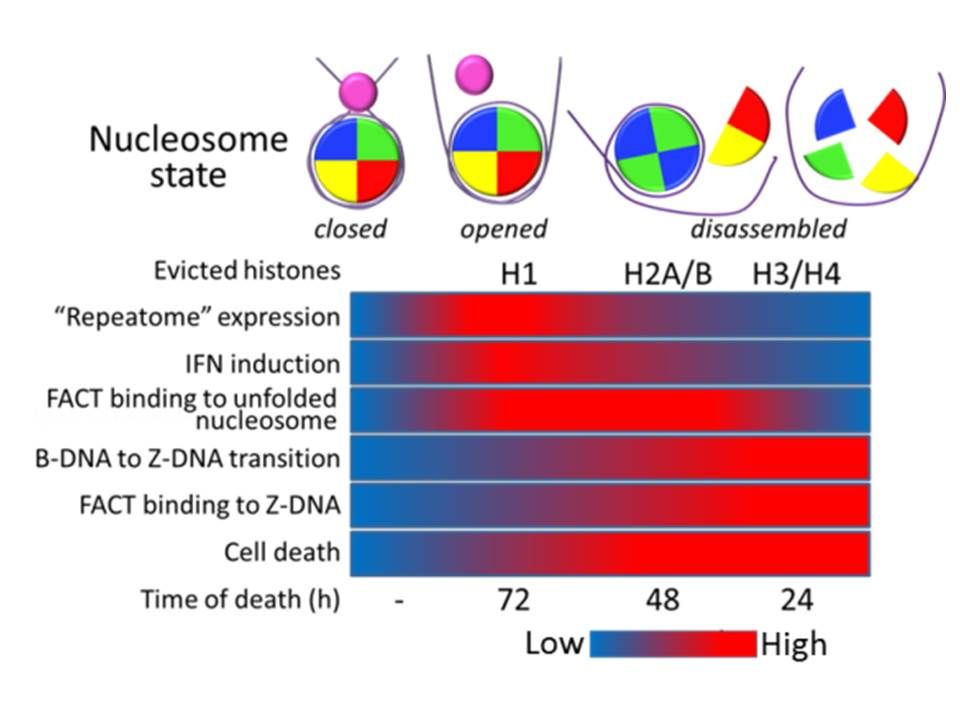
Chromatin destabilization in cells is accompanied by de-silencing of heterochromatin, accumulation of double stranded RNA due to transcription of repetitive heterochromatic elements and induction of type I interferon response.
Since interferon may induce apoptosis, this mechanism may be responsible for elimination of cells with destabilized chromatin.
Contact the Gurova Lab
Email: Katerina.Gurova@RoswellPark.org
Phone: 716-845-4760
Office location: Center for Genetics & Pharmacology (CGP) L3-301A
Lab Location: Center for Genetics & Pharmacology (CGP) L3-121 & 122
Department of Cell Stress Biology
Roswell Park Comprehensive Cancer Center
Elm and Carlton Streets
Buffalo, NY 14263