What we do
The Genomics Shared Resource (GSR) at Roswell Park Comprehensive Cancer Center offers sample-to-data services, with an expert technical staff performing all aspects of sample preparation, QC, assay design and analysis.
The GSR is a broadly functioning core that provides an integrated set of tools and services for genomic analysis. Our long-standing history and contribution to the Human Genome Program through clone generation, high throughput mapping, array technology development and distribution of these resources worldwide provides a track record documenting our expertise.
The suite of technology and platform offerings virtually encompasses all available genomics platforms, allowing tremendous flexibility and complete research solutions including full-service sample to data offerings, training and education on instruments and methodologies, complete discovery/screening to validation services, and full Laboratory Information Management System (LIMS) request, workflow and sample management. The GSR currently house state-of-the-art Illumina NovaSeq 6000 and NextSeq500 sequencers necessary for carrying out large scale Next Generation Sequencing (NGS) projects. We have extensive collaborations with Roswell Park and external investigators carrying out projects using NGS as well as other companion technologies such as 10x Genomics, Nanostring nCounter, Illumina iScan and Sanger sequencing.
Technology
Illumina NovaSeq 6000
Illumina NextSeq 500
Illumina MiSeq
Illumina iScan
10x Genomics Chromium X and Chromium Controller
Other equipment
- Caliper Sciclone NGS workstation
- Opentron OT-2
- Covaris E-220
- ABI QuantStudio 6
- ABI 3500 Genetic Analyzer
- Nanostring nCounter
- Agilent Bioanalyzer 2100
- Agilent TapeStation 4200
- Qubit 3 Fluorometer
- SpectraMax® Gemini™ XPS Microplate Spectrofluorometer
- Evos FL Auto 2 microscope
- Eppendorf Vacufuge plus
Nucleic acid extraction & QC
- DNA/RNA extraction
- Tissue, FFPE, Cells, blood, plasma (cfDNA), saliva, buccal swabs etc.
- Microbiome
- Feces, tissue and saliva
Bioanalyzer
TapeStation
Qubit
SpectraMax
Covaris
Sequencing
- Next generation sequencing
- RNA-seq
- Whole Genome (WGS)
- Whole Exome (WES)
- Methylation (WGBS, RRBS)
- Metagenome (16s, meta-GEX, meta-genome)
- EpiGen: ChIP, ATAC, cut&run
- Sanger sequencing
- Sanger sequencing
- Fragment analysis
NovaSeq
ABI 3500
NextSeq
MiSeq
Single cell and spatial profiling
- Single cell transcriptomics
- Gene Expression Profiling
- CRISPR Screening
- Gene & Cell Surface Protein
- Immune Profiling
- Single cell epigenomics
- Chromatin Accessibility (ATAC-seq)
- Multiome - GEX+ATAC-seq
- Spatial transcriptomics
- 10x Visium
Chromium X
10X Visium
Companion technologies
- Illumina iScan
- SNP arrays
- Methylation arrays
- NanoString nCounter
- Gene expression panels
- CNV
- Real time PCR
- TaqMan: GEX, SNP
- SYBR GEX
iScan
nCounter
QuantStudio 6 Flex
Services
The Genomics Shared Resource prepares RNA and DNA from a variety of sample types, using several extraction protocols depending on the sample size and downstream application.
- DNA (cells, tissues, blood, FFPE): Qiagen DNeasy, PAXgene Blood DNA, Covaris truXTRAC FFPE total NA
- RNA (cells, tissues, blood, FFPE): TRIzol, Qiagen miRNeasy, PAXgene Blood miRNA, Covaris truXTRAC FFPE total NA
- miRNA enriched: Qiagen miRNeasy
- Sample processing for microbiome analyses: DNeasy PowerSoil Pro, ZymoBIOMICS DNA
Nucleic acid quality control
- Qubit and SpectraMax fluorescent plate reader: picogreen, ribogreen quantitation
- Bioanalyzer 2100: RNA and DNA chips
- Agilent Tapestation 4200
Sample requirements
- Cell pellets: 250,000 to 1 million cells. Snap frozen or fresh cell pellet
- Tissue: 10 to 25mg frozen tissue
- Blood: Paxgene Tubes for RNA, 300ul of whole blood for plate format extraction
- FFPE: 10 slides or curls, 5 to 10um thickness
Contact Prashant Singh at 716-845-3869, Prashant.Singh@RoswellPark.org for projects specific sample requirements.
The GSR provides a full complement of NGS services, including Whole Genome Sequencing (WGS), Whole Exome-seq (WES), Whole transcriptome (RNA-seq and small RNA-seq), WGBis-seq (Methyl-seq), ChIP-Seq, ATAC-seq, and targeted DNA sequencing. Additionally, GSR also provides single-cell sequencing and spatial transcriptomics using 10x Genomics Chromium X and 10x Genomics Visium platforms, respectively, which enables detailed analysis of tissue heterogeneity at a single cell and whole tissue level. The GSR houses state-of-the-art Illumina NovaSeq 6000, NextSeq500 and MiSeq sequencers necessary for carrying out Next Generation Sequencing (NGS) projects. Furthermore, the facility contains a cyclone robot for automation of large-scale projects.
Genomics
Sequencing DNA of genomic DNA is used for interrogation and identification of DNA mutations, SNPs, Copy Number Variations (CNVs) and structural variations (SVs). The GSR provides different types of DNA sequencing services including Whole Genome sequencing, Whole Exome Sequencing and targeted sequencing.
| Library Type | Library Method/Kits | Sample Input/Quality | Application |
|---|---|---|---|
| Whole Genome Sequencing (WGS) | Illumina DNA PCR-Free, KAPA Hyperprep and others | 1ng to 2ug | Whole genome sequencing for DNA mutation, SNP, CNV and structural variation analysis. |
| Whole Exome Sequencing (WES) | Agilent SureSelect XT HS2 DNA | 10ng to 200ng | Sequencing of all exons. Used for somatic mutation analysis, small in/dels and SNP. SureSelect Human All Exon V8, 35.1 Mb target region with an efficient end-to-end design size of 41.6 Mb. Also available for other species such as mouse, rat and dog. |
| Targeted DNA sequencing | Custom amplicon, Agilent SureSelect XT HS2 DNA and IDT rhAmpSeq | 10ng to 1ug | Sequencing of specific DNA targets ( one mutation to custom targets of several Mbs) |
Transcriptomics
Transcriptome sequencing (RNA-seq) provides a comprehensive measurement of levels of transcripts and their isoforms at a given biological moment and used to identify differentially expressed genes between different samples. The GSR provides several types of RNA-seq services suitable for different samples types, RNA inputs, RNA quality, and different RNA species such as mRNA, smallRNA, noncodingRNA, or microRNAs.
| Library Type | Library Method/Kits | Sample Input/Quality | Application |
|---|---|---|---|
| Total RNA-seq | KAPA RNA HyperPrep Kit with RiboErase (HMR). Ribosomal Depletion. | 25 ng – 1 µg, RIN>4 | Whole Transcriptome sequencing. Stranded library. Gene expression analysis, detection of gene fusions, including noncoding transcripts, isoforms and novel transcript identification. |
| mRNA-seq | KAPA mRNA HyperPrep Kit. poly(A) enrichment | 50 ng – 1 µg, RIN>7 | Whole Transcriptome sequencing. Stranded library, Gene expression analysis of protein coding genes and detection of gene fusions. |
| FFPE whole Transcriptome | SureSelect XT HS2 RNA | 10 - 200 ng of total RNA from intact or highly fragmented FFPE samples (DV200>20%) | Gene expression analysis using exon capture with SureSelect Human All Exon V8+UTR kit. Suitable for highly degraded RNA such as from FFPE samples. |
| Pico-input | Takara/Clotech SMARTer Stranded Total RNA-Seq Kit - Pico Input (Ribo depletion) | 250 pg–10 ng, RIN>4 | Whole Transcriptome sequencing with low input samples. Stranded library. |
| Ultra Low input | SMART-Seq v4 Kit ultra-low input (poly(A) enrichment) | Up to 1,000 cells or 10 pg–10 ng Total RNA, RIN>7 | Whole Transcriptome sequencing with ultra low input samples. Unstranded library. |
| Small RNA | SMARTer smRNA-Seq Kit | 1 ng–2 µg, RIN>7 | Expression analysis of small non-coding RNAs (smRNAs) including microRNAs |
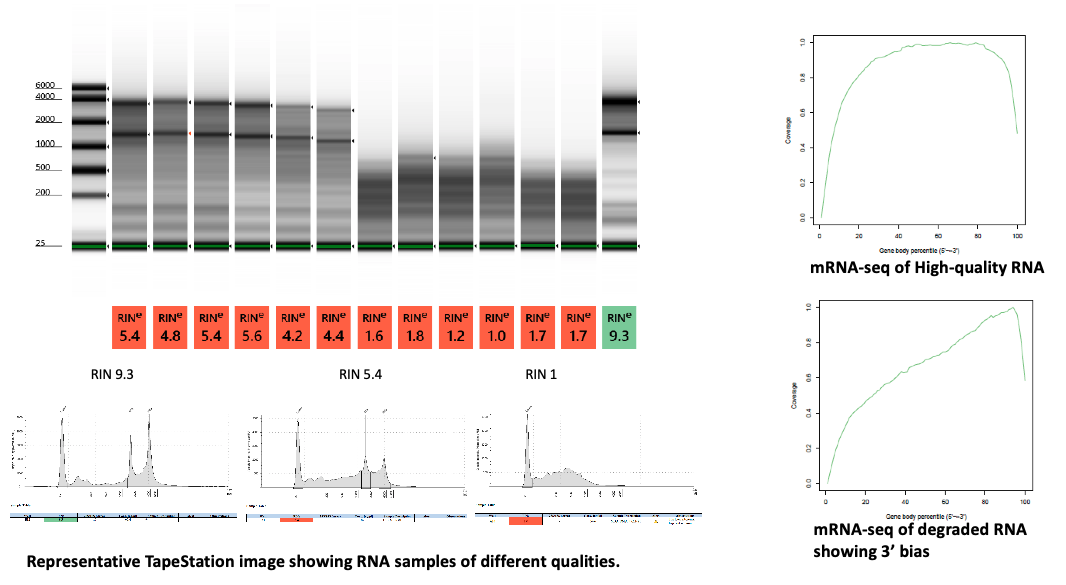
It is critical to treat RNA samples with DNase for RNA-seq applications. The qualitative assessment of RNA samples is performed using Agilent Bioanalyzer or TapeStation and RNA samples with RIN >7 are considered high quality and suitable for all the assays. Degraded RNA samples can be analyzed (i.e. paraffin, formalin fixed tissues) with appropriate method. Below examples show different RNA quality samples. Use of mRNA-seq is not recommended for lower quality samples as it will result in 3’ bias.
Epigenomics
Epigenomics approaches are used to the study of changes in gene activity caused by mechanisms other than DNA sequence changes such as DNA methylation, smallRNA–mediated gene regulation, DNA–protein interactions, histone modification, chromatin accessibility and more. The GSR provides various epigenomics assays utilizing NGS and microarray platforms.
| Library Type | Library Method/Kits | Sample Input/Quality | Application |
|---|---|---|---|
| Whole Genome BiSulfite Sequencing (WGBS) | xGen™ Methyl-Seq Lib Prep | 10 to 100ng | Genome wide CpG methylation analysis |
| Reduced Representation Bisulfite Sequencing (RRBS) | Ovation® RRBS Methyl-Seq | 10 to 100ng | Genome-wide coverage of CpGs in islands at single-base resolution, measures 10-15% of all CpGs in genome. Reduced sequencing requirement and cost. |
| Targeted methylation Sequencing | Amplicon Bases | 10 to 100ng | Targeted methylation of 1 to 20 CpGs |
| ChIP-seq | Takara/Clotech ThruPLEX | 1 to 100ng | Transcription factor and histone modifications. Users need to submit IP DNA |
| Cut&Run | Takara/Clotech ThruPLEX | 1 to 100ng | Transcription factor and histone modifications. Users need to submit IP DNA |
| ATAC-seq | Custom | Use provided library | Chromatin accessibility. Users need to submit tagmented DNA. |
Metagenomics
Metagenomics allows characterization of composition and variation of microbial community in any given sample. The GSR provides full service for metagenomics including DNA RNA extraction and various NGS approaches.
| Library Type | Library Method/Kits | Sample Input/Quality | Application |
|---|---|---|---|
| Metagenomics-16S ribosomal RNA gene sequencing | Amplicon based, V3-V4 regions | 10-100ng DNA | Phylogenetic classifications such as genus or species of diverse bacteria or archaea. |
| Metagenomics-ITS (Internal Transcribed Spacer) | Amplicon based, ITS1 region | 10-100ng DNA | Phylogenetic analysis of different fungi |
| Meta-genome | Shotgun sequencing ( Illumina DNA PCR free or Nextera XT kit) | 1ng to 2ug DNA | Profiling of diverse microorganisms such as bacteria, fungi, and viruses as we well profile of microbial genes. |
In recent years, single cell sequencing has provided novel insights into tissue and tumor microenvironment heterogeneity at single-cell resolution. This knowledge has helped identify novel cell types and opened avenues for innovative, therapeutic interventions. The GSR provides single cell sequencing application using 10x Genomics Chromium X platform.
Single cell applications supported by the GSR
- Single cell transcriptomics
- Gene Expression Profiling (3’ and 5’ gene expression)
- Gene Expression CRISPR Screening
- Gene & Cell Surface Protein
- Immune Profiling & Antigen Specificity
- Single cell epigenomics
- Chromatin Accessibility (ATAC-seq)
- Multiome: Gene expression+ATAC-seq
Information on sample preparation for submission can be found here.
Sample Requirement:
- Cells must be in a single cell suspension and free of debris and cell aggregates.
- Cell viability should be >70%, lower viability may produce compromised results
- Cell stock should be between 500-1200 cells/ul in 1.5ml Eppendorf tubes, volume 30-50ul
- Cells should be re-suspended in PBS+0.04% BSA or complete media,
- Sample should be delivered on ice with completed sample submission form by 2pm.
- Please provide information about desired target cell recovery (100-20000 cells/sample) and assay type (3’GEX, 5’GEX, ATACseq, TCR/BCR)
How many cells to submit?
10x platform has a capture rate of ~60% so for example if desired target cell recovery is 10,000 cells, then 16,000 cells are loaded into the system. The chart provides information about number of cells required for different target cell recoveries. We also need 10ul cell suspension for QC.
A practical example:
Cell concentration: 600 cells/ul
Target cell recovery: 5,000 cells
In this case we will need 10ul of cell suspension for counting and QC (6,000 cells) and 13.8 ul of cell suspension will be loaded into the Chromium system (8,280 cells). We will need minimum 23.8ul cell suspension (14,280 cells). We request at least double the amount of minimum required to make sure we have enough cell to re-load if there was an issue with chromium run. For example, in this case we will request 50ul cell suspension or 30,000 cells.
Spatial profiling:
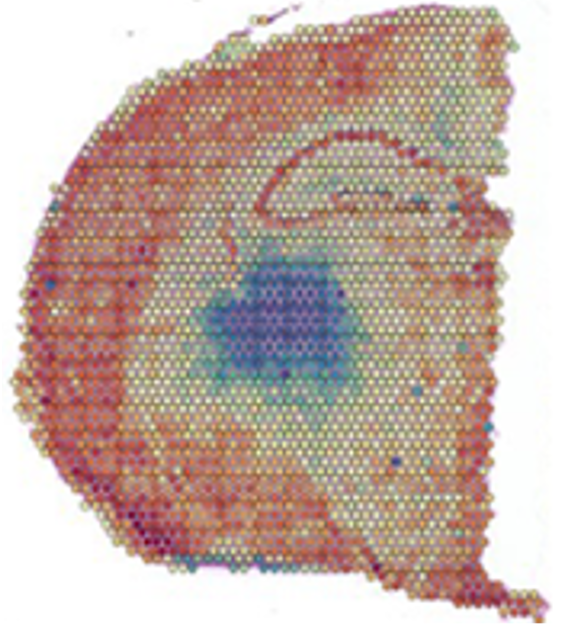
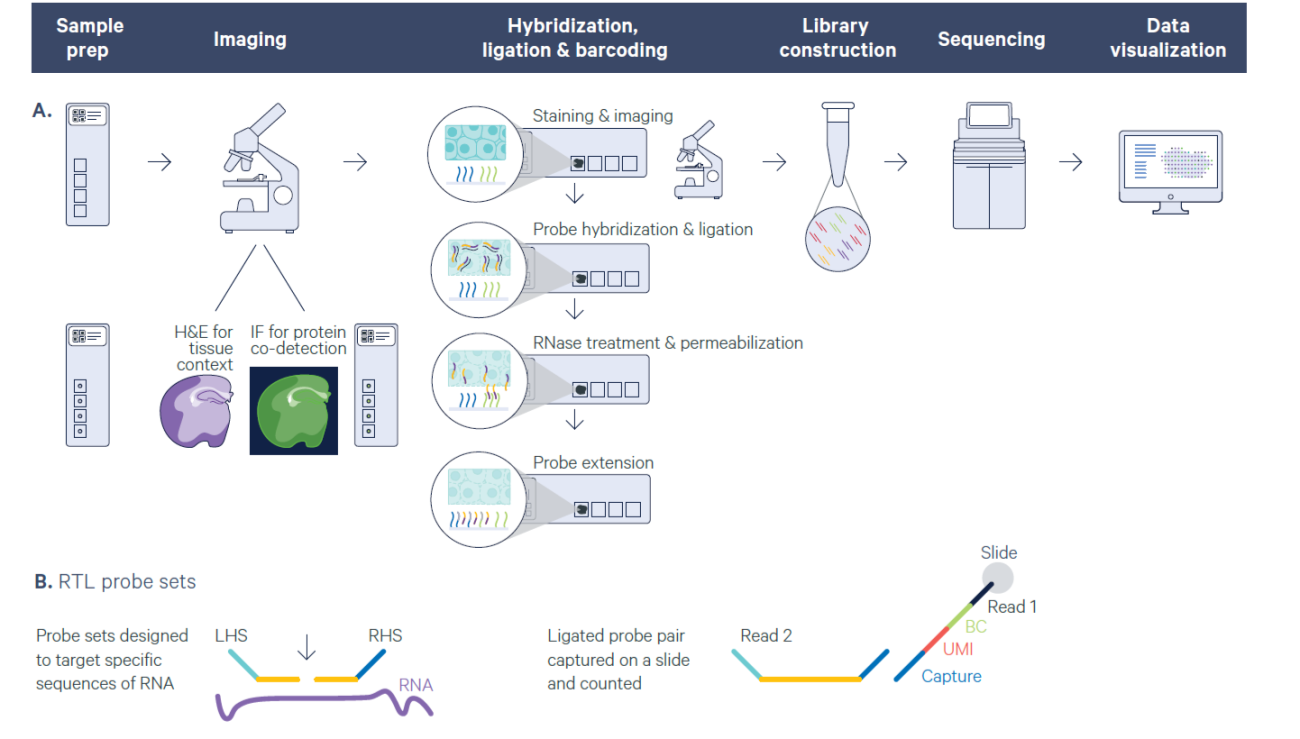
Single cell sequencing provides comprehensive analysis of tissue heterogeneity and identification of different cells types; however, spatial information of cells withing tissue is lost due to tissue dissociation. Spatial profiling approaches enables analysis of relationship between cells and their microenvironment within tissue context. Spatial transcriptomics is a groundbreaking molecular profiling method that provides deep understanding of normal development and disease pathology by measuring gene expression within a tissue context. The GSR utilized 10x Genomics Visium platform to perform spatial transcriptomics. The Visium platform can be used for both fresh frozen and FFPE tissue samples.
The Visium is a complex work involving the genomics and pathology expertise. Please contact Dr. Prashant Singh to discuss details about sample requirement and experimental design.
The Genomics Shared Resource provides genome wide SNP, Copy Number Variation (CNV) and DNA methylation services using microarray technology (also known as BeadArray technology).
Methylation arrays
We use methylation arrays for both human and mouse-derived samples.
Human methylation array
Genome-wide methylation analysis using Illumina's Infinium MethylationEPIC BeadChip requires samples to be run in batches of 16. We require 1ug of high molecular weight DNA (via picogreen quantitation) per sample.
Illumina's MethylationEpic BeadChip contains more than 850K of highly informative CpG sites covering 99% of RefSeq genes, and 95% of all known CpG islands, along with enhancer sits and other content categories. The array encompasses CpG sites outside of CpG islands, Non-CpG methylated sites, differentially methylated sites identified in tumor versus normal, FANTOM 5 enhancers, ENCODE open chromatin and enhancers, miRNA promoters and DNase hypersensitive sites. The MethylationEpic array contains more than 90% of the CpGs on the Illumina Methylatio450 plus an additional 350,000 CpGs in enhancer regions.
Mouse methylation array
Genome-wide methylation analysis of mouse samples is performed using Infinium Mouse Methylation BeadChip. This array interrogates more than 285k methylation sites selected to provide coverage of CpG islands, translation start sites, enhancers, imprinted loci, and other regions. This array requires samples to be run in batches of 24. We require 1ug of high molecular weight DNA (via picogreen quantitation) per sample.
SNP arrays
The GSR provides global genotyping analyses, DNA copy number variation (CNV) and loss of heterozygosity (LOH) services using the Illumina Infinium products. The Illumina BeadChips provide a broad spectrum of whole-genome DNA Analysis products to support a variety of experimental designs. Researchers have the flexibility to use panels of 300,000 to nearly 5,000,000 markers per sample, depending on their study goals. All BeadChips provide powerful and integrated genome-wide SNP genotyping, LOH and CNV detection.
Sample requirements
We request 1ug of high-quality genomic DNA for Illumina arrays. Input requirement for methylation arrays is 500ng and 200ng for SNP arrays. Most standard genomic DNA isolation protocols will yield DNA of high enough purity for microarray analysis. It is highly recommended to check genomic DNA for degradation and RNA contamination. It is critical to use purified and accurate amounts of DNA for fluorescent labeling. Degraded DNA from FFPE samples can be used with Illumina arrays after performing DNA restoration. Please contact Prashant Singh at 716-845-3869 for more details.
Disclaimer for SNP data
These assays are intended solely for research purposes. The results provided in this report are not certified by the New York State Department of Health for or intended for use in clinical diagnostic testing or treatment. In using these assay results, the user agrees that Roswell Park Comprehensive Cancer Center shall have no liability for claims by, or damages of any kind whatsoever to, a user of these assay results for a decision or action taken in reliance on the information provided in the results, including any direct, indirect, special, incidental or consequential damages relating to its use.
Roswell Park makes no warranties of any kind, express or implied, as to the merchantability or fitness for a particular use of the assay results. These assay results are provided "as is" and without warranty that their use will not infringe any patent or other proprietary right of a third party.
The Genomics Shared Resource provides gene expression analysis on several different platforms depending on the organism, starting material and size of your project.
RNA sequencing (RNA-Seq)
The GSR uses several types of RNA-seq library prep methods to perform global gene expression analyses depending on sample and analysis requirement. More details are provided in NGS section.
NanoString nCounter gene expression
The NanoString nCounter Analysis System uses a digital technology to directly and simultaneously, measure multiple genomic targets in tissue samples, without the need for amplification or library preparation. The GSR utilizes the nCounter Analysis System for analysis of NanoString gene panels from, custom gene panels and microRNA expression. NanoString gene expression panels are available for both human and mouse.
Quantitative Realtime PCR (qPCR)
The GSR provides full service for qPCR analysis using QuantStudio 6 Flex Real-Time PCR System. The qPCR is used for gene expression analysis of small number of genes (1 to 20 genes). We can run both TaqMan as well as SYBR green based assays. The qPCR is used for expression analysis of mRNA as well as microRNAs.
Sample requirement
Generally, 100ng to 1 µg total RNA is required, depending on the assay.
Note: It is critical to treat RNA samples with DNase for RNA-seq applications. The qualitative assessment of RNA samples is performed using Agilent Bioanalyzer or TapeStation and RNA samples with RIN >7 are considered high quality and suitable for all the assays. Degraded RNA samples can be analyzed (i.e. paraffin, formalin fixed tissues) with NanoString and RNA-seq.
The Genomics Shared Resource provides full service of Sanger sequencing and fragment analysis. The GSR houses two ABI 3500 Genetic Analyzer for Sanger sequencing and fragment analysis.
Online ordering
All orders must be submitted using LIMS system. With our web-based ordering system, you will be able to submit your orders electronically and retrieve your sequence data when completed.
Contact us
Khin Marlar
Genomics Shared Resource
DNA Sequencing Laboratory
Center for Genetics and Pharmacology, L1-104
Roswell Park Comprehensive Cancer Center
Elm and Carlton Streets
Buffalo, NY 14263
Email: Khin.Marlar@RoswellPark.org
Phone: 716-845-8314
Fax: 716-845-1579
When mailing samples, it is preferable to choose overnight delivery. While DNA samples are stable at ambient temperature, prolonged stays at these temperatures can lead to degradation of DNA and subsequent deterioration of sequence quality. Information on the primers we provide can be found below.
Fee schedule
All Roswell Park CCSG member and nonmember prices are available by contacting Prashant Singh at 716-845-3869 or Prashant.Singh@RoswellPark.org.
Sanger sequencing legal disclaimer
The DNA sequencing services provided by Roswell Park’s Genomic Shared Resource are intended solely for research purposes. These DNA sequencing services are not intended or certified by the New York State Department of Health for use in clinical diagnostic testing or treatment, including but not limited to patient education.
Roswell Park Comprehensive Cancer Center shall have no liability for claims by, or damages of any kind whatsoever to, a user of these DNA sequencing services for a decision or action taken in reliance on the information provided in the DNA sequencing. Such damages include, without limitation, direct, indirect, special, incidental or consequential damages.
The Genomics Shared Resource provides Human Cell Line Authentication service using short tandem repeats (STR) profiling.
Background
The cell lines are important research tools for basic biomedical and pre-clinical research. However, contamination and misidentification of cell lines is a major problem leading to spurious results and irreproducibility of data. Recent changes in NIH grant submission guidelines require all cell lines to be authenticated by chromosomal analysis or STR profiling.
STR profiling
The cell line authentication is performed using STR loci which are also used in DNA fingerprinting. The STR markers are polymorphic DNA loci that contain repeated nucleotide sequence and the number of repeats varies for each individual, combination of repeats are used to match cell lines with their reported profile (ATCC, DSMZ, etc).
We use AmpFlSTR Profiler PCR Amplification Kit from Applied Biosystems which amplifies 15 STR loci and the amelogenin gene in one single multiplex PCR reaction. The amplification products are fluorescently labeled and analyzed using the ABI 3130xl sequencer. Each peak in the electropherogram represents an allele and by comparison of the sample data to an allelic ladder, the number of repeat units is determined. This numeric value for each repeat is then compared to cell line STR profile reported in ATCC and DSMZ databases to confirm the cell line.
Fee schedule
All Roswell Park CCSG member and nonmember prices are available by contacting Prashant Singh at 716-845-3869 or Prashant.Singh@RoswellPark.org.
Location & hours
Roswell Park Comprehensive Cancer CenterGenomics Shared Resource
Center for Genetics and Pharmacology
Room L1-130
Elm and Carlton Streets
Buffalo, New York 14263
Monday – Friday, 8 a.m. – 5 p.m.
This shared resource is funded by NCI P30CA16056. Publications should cite the core grant in the acknowledgment section, if publications use data generated by the shared resource.
An example: This work was supported by National Cancer Institute (NCI) grant P30CA016056 involving the use of Roswell Park Comprehensive Cancer Center’s Genomic Shared Resource.
Genomics Shared Resource
The Genomics Shared Resource (GSR) offers sample-to-data services, with expert technical staff performing all aspects of sample preparation, QC, assay design, and analysis. The mission of the GSR is to provide state-of-the-art instrumentation and expertise that enables its users to acquire and analyze genomic data sets across basic, translational, clinical, and population studies. Our long-standing history and contribution to the Human Genome Program through clone generation, high-throughput mapping, array technology development, and distribution of these resources worldwide provides a track record documenting our expertise that can be extended to the outside community. The GSR offers investigators a full spectrum of services, including Next Generation Sequencing (NGS), genotyping (targeted and global), methylation (targeted and global), copy number and expression analysis (gene and miRNA), single-cell sequencing, and Sanger sequencing. The facility houses an Illumina iScan system, two Applied Biosystems QuantStudio6 Real-Time PCR Systems, two Applied Biosystems 3500 Genetic Analyzers, Caliper Sciclone NGS workstation, 10x Genomics Chromium X system and Nanostring nCounter systems, as well as Illumina (NovaSeq 6000, NextSeq 500, and MiSeq) sequencers. The GSR provides a full complement of NGS services, including Whole Genome Sequencing (WGS), Whole Exome-seq (WES), Whole transcriptome (RNA-seq and small RNA-seq), WGBis-seq (Methyl-seq), ChIP-Seq, and targeted DNA sequencing. Additionally, GSR also provides single-cell sequencing and spatial transcriptomics using 10x Chromium X and Visium platforms, respectively, which enables detailed analysis of tissue heterogeneity at a single cell and whole tissue level. The GSR also can perform methylation analysis of intact or compromised DNA samples using the Illumina Infinium MethylationEPIC BeadChip (global) as well as full-service SNP/CNV (300K-4.5M SNPs) genotyping using Illumina BeadChip technology. The GSR provides many levels of customizable validation technologies to fit any sized project, including custom Illumina BeadChips, qPCR, nCounter digital detection assays, and Sanger sequencing. The GSR is also a repository for the RP11 and RP23 BAC genomic clone libraries, which are available for clone selection, characterization, FISH mapping, and distribution. The GSR is a state-of-the-art facility that utilizes a variety of platforms to accommodate any size request and monitors all processes using a Laboratory Information Management Systems (LIMS), which tracks online requests, sample submission, workflow, and sample processing to ensure that quality reproducible data are generated efficiently. The GSR operates as a CLIA-compliant space, which allows it to perform correlative studies related to clinical trials as well as pharma projects.
At an institutional level, the GSR interacts with other shared resources such as the Biostatistics and Bioinformatics shared resources, DBBR, and Pathology Network Shared Resource (PNSR) to facilitate centralized high-throughput sample preparation from archival and prospective cohorts to better manage investigator-initiated projects and research questions.
Publications
- Alam A, Levanduski E, Denz P, Villavicencio HS, Bhatta M, Alhorebi L, Zhang Y, Gomez EC, Morreale B, Senchanthisai S, Li J, Turowski SG, Sexton S, Sait SJ, Singh PK, Wang J, Maitra A, Kalinski P, DePinho RA, Wang H, Liao W, Abrams SI, Segal BH, Dey P. Fungal mycobiome drives IL-33 secretion and type 2 immunity in pancreatic cancer. Cancer Cell. 2022 Feb 14;40(2):153-167
- M Maximilian, Merz A, Wang J, Wei L, Hu Q, Hutson N, Rondeau C, Celotto K, Belal A, Alberico R, Block AM, Mohammadpour H, Wallace P, Tario J, Luce J, Glenn GT, Singh PK, Herr M, Hahn T, Samur M, Munshi M, Liu S, McCarthy P, Hillengass J. Deciphering spatial genomic heterogeneity at a single cell resolution in multiple myeloma. Nat Commun. 2022 Feb 10;13(1):807
- Knudsen E, Kumarasamy V, Chung S, Ruiz A, Vail P, Tzetzo S, Wu J, Nambiar R, Sivinski J, Chauhan SS, Seshadri M, Abrams S, Wang J, Witkiewicz A. Targeting dual signalling pathways in concert with immune checkpoints for the treatment of pancreatic cancer. Gut 2021; 70(1): 127-138. PMC7671951
- Long MD, Jacobi JJ, Singh PK, Llimos G, Wani SA, Rowsam AM, Rosario SR, Hoogstraat M, Linder S, Kirk J, Affronti HC, Bergman A, Zwart W, Campbell MJ, Smiraglia DJ. Reduced NCOR2 expression accelerates androgen deprivation therapy failure in prostate cancer. Cell Rep. 2021 Dec 14;37(11):110109.
- Wei L, Christensen SR, Fitzgerald M, Graham J, Hutson ND, Zhang C, Huang Z, Hu Q, Zhan F, Xie J, Zhang J, Liu S, Remenyik E, Gellen E, Colegio OR, Bax M, Xu J, Lin H, Huss W, Foster B, Paragh G. Ultradeep sequencing differentiates patterns of skin clonal mutations associated with sun-exposure status and skin cancer burden. Sci Adv 2021; 7(1): eabd7703. PMC7775785
- Want MY, Tsuji T, Singh P, Thorne JL, Matsuzaki J, Karasik E, Gillard B, Cortes E, Koya R, Lugade A, Odunsi A, Battaglia S. WHSC1/NSD2 regulates immune infiltration in prostate cancer. J Immunother Cancer 2021; 9(2): e001374. PMC7887377
- Oba T, Long M, Keler T, Marsh HC, Minderman H, Abrams S, Liu S, Ito F. Overcoming primary and acquired resistance to anti-PD-L1 therapy by induction and activation of tumor-residing cDC1s. Nat Commun 2020; 11(1): 5415. PMC7592056
- Yang Y, Jain RK, Glenn S, Xu B, Singh P, Wei L, Hu Q, Long M, Hutson N, Wang J, Battaglia S, George S. Complete response to anti-PD-L1 antibody in a metastatic bladder cancer associated with novel MSH4 mutation and microsatellite instability. J Immunother Cancer 2020; 8(1): e000128. PMC7206971
- Ku SY, Rosario S, Wang Y, Mu P, Seshadri M, Goodrich ZW, Goodrich MM, Labbe DP, Cortes E, Wang J, Long HW, Xu B, Brown M, Loda M, Sawyers CL, Ellis L, Goodrich D. Rb1 and Trp53 cooperate to suppress prostate cancer lineage plasticity, metastasis, and antiandrogen resistance. Science 2017; 355(6320): 78-83. PMC5367887
- Morrison C, Liu P, Woloszynska-Read A, Zhang J, Luo W, Qin M, Bshara W, Conroy J, Sabatini L, Vedell P, Xiong D, Liu S, Wang J, Shen H, Li Y, Omilian A, Hill A, Head K, Guru K, Kunnev D, Leach R, Eng K, Darlak C, Hoeflich C, Veeranki S, Glenn S, You M, Pruitt S, Johnson C, Trump D. Whole-genome sequencing identifies genomic heterogeneity at a nucleotide and chromosomal level in bladder cancer. Proc Natl Acad Sci U S A 2014; 111(6): E672-E681. PMC3926024