What we do
The Advanced Tissue Imaging Shared Resource (ATISR) offers streamlined, multiplexed tissue analysis that helps move your project forward. We accommodate your specific needs – from tissue sectioning, staining, imaging, spatial RNA sequencing, analysis, and interpretation of your results. We work with all investigators! From researchers at Roswell Park to researchers at external academic institutions and companies in the biotech field.
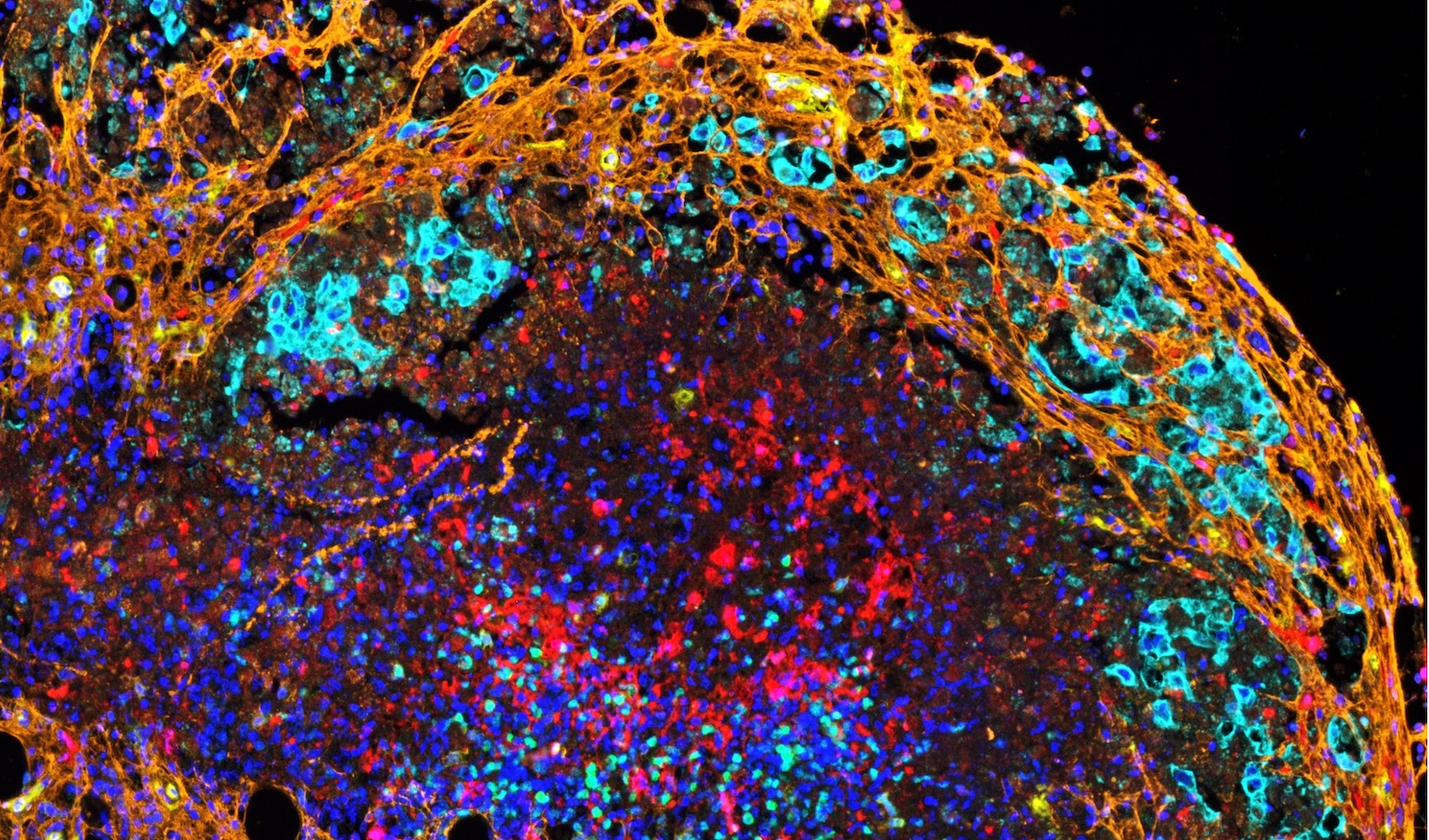
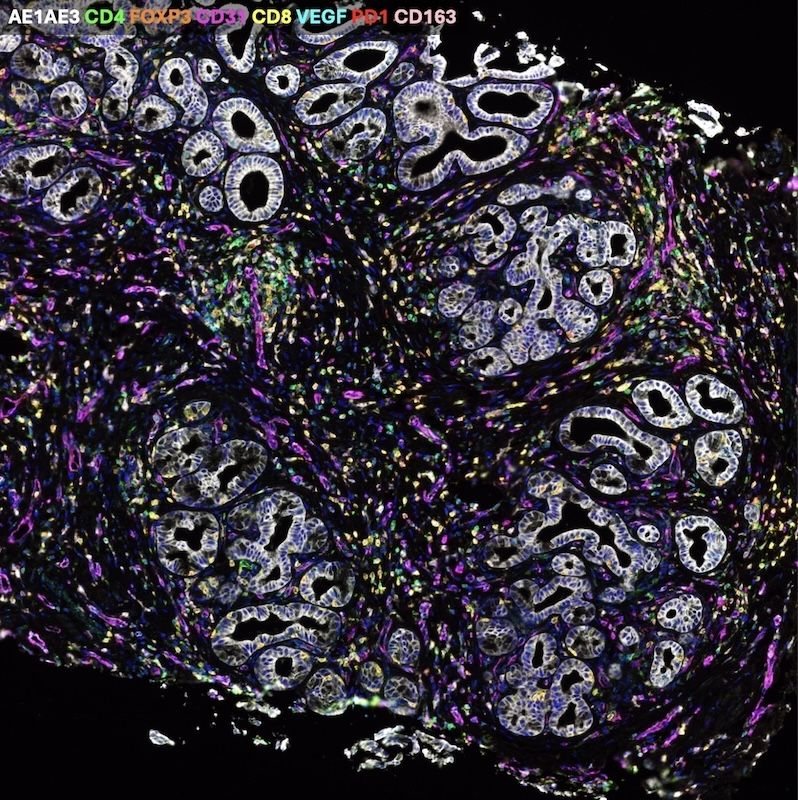
Our multispectral immunofluorescence (mIF) capabilities allow you to use up to eight biomarkers per panel on the same tissue to answer your most pressing research questions.
Learn about multispectral immunofluorescence
Our spatial transcriptomics pipeline can be performed separately or multiplexed with mIF and enables spatial whole transcriptome RNA sequencing analysis on your samples, while preserving the morphological integrity of your sample for further assessments.
If you have any questions or would like to discuss your project-specific needs, feel free to email ATISR@RoswellPark.org and we’ll promptly get back to you.
Additional services
The newly acquired high-resolution Leica DMi8 inverted microscope with the advanced Leica THUNDER technology suite, enabling 3D imaging and Computational Clearing that can produce both high magnification sub-cellular immunofluorescent images of the highest quality or crisp low magnification images of entire tumor microarrays. Image source: Leica Microsystems
The fully automated IncuCyte S3 live-cell imaging system for multi-condition cellular proliferation analysis with red/green fluorescence channels and phase contrast imaging at 4, 10, and 20X around the clock. Capable of assessing six microplates, dishes, flasks, or slides simultaneously. Image source: Sartorius Incucyte S3
2025
- Farrugia MK, Repasky EA, Chen M, Attwood K, Catalfamo K, Rosenheck H, Yao S, Mattson DM, Mukherjee S, Kukar M, Witkiewicz AK, Singh AK. Prognostic immune markers in esophageal cancer patients managed with trimodal therapy. Cancer Immunol Immunother. 2025 Jan 3;74(2):57. doi: 10.1007/s00262-024-03891-3. PMID: 39751903; PMCID: PMC11698998.
- Kumarasamy V, Wang J, Roti M, Wan Y, Dommer AP, Rosenheck H, Putta S, Trub A, Bisi J, Strum J, Roberts P, Rubin SM, Frangou C, McLean K, Witkiewicz AK, Knudsen ES. Discrete vulnerability to pharmacological CDK2 inhibition is governed by heterogeneity of the cancer cell cycle. Nat Commun. 2025 Feb 9;16(1):1476. doi: 10.1038/s41467-025-56674-4. PMID: 39924553; PMCID: PMC11808123.
- Dommer AP, Kumarasamy V, Wang J, O'Connor TN, Roti M, Mahan S, McLean K, Knudsen ES, Witkiewicz AK. Tumor Suppressors Condition Differential Responses to the Selective CDK2 Inhibitor BLU-222. Cancer Res. 2025 Feb 13. doi: 10.1158/0008-5472.CAN-24-2244. Epub ahead of print. PMID: 39945638.
- Crawford KJ, Humphrey KS, Cortes E, Wang J, Feigin ME, Witkiewicz AK, Knudsen ES, Abel EV. Targeting FGFR4 Abrogates HNF1A-driven Metastasis in Pancreatic Ductal Adenocarcinoma. bioRxiv [Preprint]. 2025 Feb 8:2025.02.06.636643. doi: 10.1101/2025.02.06.636643. PMID: 39974881; PMCID: PMC11839031.
- Wan Y, Wang J, O’Connor TN, Kumarasamy V, Sanidas I, Abrams SI, Witkiewicz AK, Knudsen ES. Intrinsic and extrinsic impacts of RB activation in Triple-Negative Breast Cancer. In submission.
- Tzetzo SL, Schultz E, Wang J, Rosenheck HR, Mahan S, Knudsen ES, Witkiewicz AK. Baseline cell cycle and immune profiles indicate CDK4/6 inhibitor response in metastatic HR+/HER2- breast cancer. In submission.
2024
- Kirk JS, Wang J, Long M, Rosario S, Tracz A, Ji Y, Kumar R, Liu X, Jamroze A, Singh PK, Puzanov I, Chatta G, Cheng Q, Huang J, Wrana JL, Lovell J, Yu H, Liu S, Shen MM, Liu T, Tang DG. Integrated single-cell analysis defines the epigenetic basis of castration-resistant prostate luminal cells. Cell Stem Cell. 2024 Aug 1;31(8):1203-1221.e7. doi: 10.1016/j.stem.2024.05.008. Epub 2024 Jun 14. PMID: 38878775; PMCID: PMC11297676.
- Wu J, Wang J, O'Connor TN, Tzetzo SL, Gurova KV, Knudsen ES, Witkiewicz AK. Separable Cell Cycle Arrest and Immune Response Elicited through Pharmacological CDK4/6 and MEK Inhibition in RASmut Disease Models. Mol Cancer Ther. 2024 Dec 3;23(12):1801-1814. doi: 10.1158/1535-7163.MCT-24-0369. PMID: 39148328; PMCID: PMC11614708.
- Kumarasamy V, Wang J, Frangou C, Wan Y, Dynka A, Rosenheck H, Dey P, Abel EV, Knudsen ES, Witkiewicz AK. The Extracellular Niche and Tumor Microenvironment Enhance KRAS Inhibitor Efficacy in Pancreatic Cancer. Cancer Res. 2024 Apr 1;84(7):1115-1132. doi: 10.1158/0008-5472.CAN-23-2504. PMID: 38294344; PMCID: PMC10982648.
- Fountzilas C, Witkiewicz A, Chatley S, Fitzpatrick V, Zonneville J, Alruwaili M, Rosenheck H, Mager D, Wang J, Krishnamurthy A, Switzer B, Attwood K, Puzanov I, Iyer R, Bakin A. YIA24-003: A Phase I Study of TAS102 Plus Talazoparib in Advanced Colorectal (CRC) and Esophagogastric (EGC) Adenocarcinomas. Journal of the National Comprehensive Cancer Network, 2024 Apr 5; 22(2.5). doi.org/10.6004/jnccn.2023.7124.
- Confer MP, Falahkheirkhah K, Surendran S, Sunny SP, Yeh K, Liu YT, Sharma I, Orr AC, Lebovic I, Magner WJ, Sigurdson SL, Aguirre A, Markiewicz MR, Suresh A, Hicks WL Jr, Birur P, Kuriakose MA, Bhargava R. Rapid and Label-Free Histopathology of Oral Lesions Using Deep Learning Applied to Optical and Infrared Spectroscopic Imaging Data. J Pers Med. 2024 Mar 13;14(3):304. doi: 10.3390/jpm14030304. PMID: 38541046; PMCID: PMC10971207.
2023
- Witkiewicz AK, Schultz E, Wang J, Hamilton D, Levine E, O'Connor T, Knudsen ES. Determinants of response to CDK4/6 inhibitors in the real-world setting. NPJ Precis Oncol. 2023 Sep 13;7(1):90. doi: 10.1038/s41698-023-00438-0. PMID: 37704753; PMCID: PMC10499925.
- Gandhi S, Opyrchal M, Grimm MJ, Slomba RT, Kokolus KM, Witkiewicz A, Attwood K, Groman A, Williams L, Tarquini ML, Wallace PK, Soh KT, Minderman H, Maguire O, O'Connor TL, Early AP, Levine EG, Kalinski P. Systemic infusion of TLR3-ligand and IFN-α in patients with breast cancer reprograms local tumor microenvironments for selective CTL influx. J Immunother Cancer. 2023 Nov;11(11):e007381. doi: 10.1136/jitc-2023-007381. PMID: 37963636; PMCID: PMC10649898.
- Mohammadpour H, Tsuji T, MacDonald CR, Sarow JL, Rosenheck H, Daneshmandi S, Choi JE, Qiu J, Matsuzaki J, Witkiewicz AK, Attwood K, Blazar BR, Odunsi K, Repasky EA, McCarthy PL. Galectin-3 expression in donor T cells reduces GvHD severity and lethality after allogeneic hematopoietic cell transplantation. Cell Rep. 2023 Mar 28;42(3):112250. doi: 10.1016/j.celrep.2023.112250. Epub 2023 Mar 15. PMID: 36924493; PMCID: PMC10116561.
- Cornwell AC, Tisdale AA, Venkat S, Maraszek KE, Alahmari AA, George A, Attwood K, George M, Rempinski D, Franco-Barraza J, Seshadri M, Parker MD, Cortes Gomez E, Fountzilas C, Cukierman E, Steele NG, Feigin ME. Lorazepam Stimulates IL6 Production and Is Associated with Poor Survival Outcomes in Pancreatic Cancer. Clin Cancer Res. 2023 Sep 15;29(18):3793-3812. doi: 10.1158/1078-0432.CCR-23-0547. PMID: 37587561; PMCID: PMC10502465.
2022
- Langille E, Al-Zahrani KN, Ma Z, Liang M, Uuskula-Reimand L, Espin R, Teng K, Malik A, Bergholtz H, Ghamrasni SE, Afiuni-Zadeh S, Tsai R, Alvi S, Elia A, Lü Y, Oh RH, Kozma KJ, Trcka D, Narimatsu M, Liu JC, Nguyen T, Barutcu S, Loganathan SK, Bremner R, Bader GD, Egan SE, Cescon DW, Sørlie T, Wrana JL, Jackson HW, Wilson MD, Witkiewicz AK, Knudsen ES, Pujana MA, Wahl GM, Schramek D. Loss of Epigenetic Regulation Disrupts Lineage Integrity, Induces Aberrant Alveogenesis, and Promotes Breast Cancer. Cancer Discov. 2022 Dec 2;12(12):2930-2953. doi: 10.1158/2159-8290.CD-21-0865. PMID: 36108220; PMCID: PMC9812400.
- Hamad SH, Montgomery SA, Simon JM, Bowman BM, Spainhower KB, Murphy RM, Knudsen ES, Fenton SE, Randell SH, Holt JR, Hayes DN, Witkiewicz AK, Oliver TG, Major MB, Weissman BE. TP53, CDKN2A/P16, and NFE2L2/NRF2 regulate the incidence of pure- and combined-small cell lung cancer in mice. Oncogene. 2022 Jun;41(25):3423-3432. doi: 10.1038/s41388-022-02348-0. Epub 2022 May 16. Erratum in: Oncogene. 2022 Sep;41(39):4485. doi: 10.1038/s41388-022-02442-3. PMID: 35577980; PMCID: PMC10039451.
- Knudsen ES, Kumarasamy V, Nambiar R, Pearson JD, Vail P, Rosenheck H, Wang J, Eng K, Bremner R, Schramek D, Rubin SM, Welm AL, Witkiewicz AK. CDK/cyclin dependencies define extreme cancer cell-cycle heterogeneity and collateral vulnerabilities. Cell Rep. 2022 Mar 1;38(9):110448. doi: 10.1016/j.celrep.2022.110448. PMID: 35235778; PMCID: PMC9022184.
- Kumarasamy V, Nambiar R, Wang J, Rosenheck H, Witkiewicz AK, Knudsen ES. RB loss determines selective resistance and novel vulnerabilities in ER-positive breast cancer models. Oncogene. 2022 Jul;41(27):3524-3538. doi: 10.1038/s41388-022-02362-2. Epub 2022 Jun 9. PMID: 35676324; PMCID: PMC10680093.
- Surendran S, Aboelkheir U, Tu AA, Magner WJ, Sigurdson SL, Merzianu M, Hicks WL Jr, Suresh A, Kirkwood KL, Kuriakose MA. T-Cell Infiltration and Immune Checkpoint Expression Increase in Oral Cavity Premalignant and Malignant Disorders. Biomedicines. 2022 Jul 30;10(8):1840. doi: 10.3390/biomedicines10081840. PMID: 36009387; PMCID: PMC9404942.
2021
- Pearson JD, Huang K, Pacal M, McCurdy SR, Lu S, Aubry A, Yu T, Wadosky KM, Zhang L, Wang T, Gregorieff A, Ahmad M, Dimaras H, Langille E, Cole SPC, Monnier PP, Lok BH, Tsao MS, Akeno N, Schramek D, Wikenheiser-Brokamp KA, Knudsen ES, Witkiewicz AK, Wrana JL, Goodrich DW, Bremner R. Binary pan-cancer classes with distinct vulnerabilities defined by pro- or anti-cancer YAP/TEAD activity. Cancer Cell. 2021 Aug 9;39(8):1115-1134.e12. doi: 10.1016/j.ccell.2021.06.016. Epub 2021 Jul 21. PMID: 34270926; PMCID: PMC8981970.
- Fountzilas C, Bajor DL, Mukherjee S, Saltzman J, Witkiewicz AK, Maguire O, Minderman H, Nambiar R, Rosenheck HR, Knudsen ES, Muhitch JB, Abrams SI, Wang C, Hutson AD, Attwood K, Hicks KA, Jurcevic JA, Kalinski P, Iyer R, Boland PM. Phase Ib/II Study of Cetuximab plus Pembrolizumab in Patients with Advanced RAS Wild-Type Colorectal Cancer. Clin Cancer Res. 2021 Dec 15;27(24):6726-6736. doi: 10.1158/1078-0432.CCR-21-1650. Epub 2021 Oct 13. PMID: 34645646; PMCID: PMC10234430.
- Kumarasamy V, Vail P, Nambiar R, Witkiewicz AK, Knudsen ES. Functional Determinants of Cell Cycle Plasticity and Sensitivity to CDK4/6 Inhibition. Cancer Res. 2021 Mar 1;81(5):1347-1360. doi: 10.1158/0008-5472.CAN-20-2275. Epub 2020 Dec 15. PMID: 33323381; PMCID: PMC8026500.
- Gandhi S, Pandey MR, Attwood K, Ji W, Witkiewicz AK, Knudsen ES, Allen C, Tario JD, Wallace PK, Cedeno CD, Levis M, Stack S, Funchain P, Drabick JJ, Bucsek MJ, Puzanov I, Mohammadpour H, Repasky EA, Ernstoff MS. Phase I Clinical Trial of Combination Propranolol and Pembrolizumab in Locally Advanced and Metastatic Melanoma: Safety, Tolerability, and Preliminary Evidence of Antitumor Activity. Clin Cancer Res. 2021 Jan 1;27(1):87-95. doi: 10.1158/1078-0432.CCR-20-2381. Epub 2020 Oct 30. PMID: 33127652; PMCID: PMC7785669.
Knudsen ES, Kumarasamy V, Chung S, Ruiz A, Vail P, Tzetzo S, Wu J, Nambiar R, Sivinski J, Chauhan SS, Seshadri M, Abrams SI, Wang J, Witkiewicz AK. Targeting dual signalling pathways in concert with immune checkpoints for the treatment of pancreatic cancer. Gut. 2021 Jan;70(1):127-138. doi: 10.1136/gutjnl-2020-321000. Epub 2020 May 18. PMID: 32424005; PMCID: PMC7671951.
Services & fees
Project estimates are available upon request. Please complete the Initial Inquiry Form and email to ATISR@RoswellPark.org.
Meet our team
Jason Wang, PhD
HRI Scientist
Phone: 716-845-1300, ext. 2244
Email: Jianxin.Wang@RoswellPark.org
William Molini, BS/MS
Research Technologist
Phone: 716-845-1300, ext. 6035
Email: William.Molini@RoswellPark.org
Thomas N. O’Connor, PhD
HRI Scientist
Phone: 716-845-1300, ext. 5096
Email: Thomas.O’Connor@RoswellPark.org
Location and hours
Roswell Park Comprehensive Cancer Center
Advanced Tissue Imaging Shared Resource
Medical Research Center, Room 109
Elm and Carlton Streets
Buffalo, New York 14263
Monday - Friday, 7:30 a.m. - 5 p.m.
This shared resource is funded by NCI P30CA16056. Publications should cite the core grant in the acknowledgment section if publications use data generated by the shared resource.