What we do
The Biorepository & Laboratory Services (BLS) shared resource at Roswell Park Comprehensive Cancer Center is a comprehensive data and biospecimen bank (malignant and normal tissue and their derivatives, blood and bone marrow) for research along with a range of services that accompany biospecimens. The overall mission of BLS is to facilitate access to human tissue, blood and bone marrow for investigators with Institutional Review Board (IRB) approved protocols emphasizing translational research. BLS plays an essential role at Roswell Park, helping investigators to establish Biospecimen and Data Research (BDR) protocols and supporting the submission of grants by providing feasibility analysis, cost estimates and letters of support.
All biospecimens can be linked with clinical data from the Biomedical Research Informatics Shared Resource (BRISR) and Shared Resource Data Managers.
Functionally, the BLS is divided into:
- Biospecimens, procurement and banking
- Genomic DNA
- Bone marrow mononuclear cells (BMMC)
- Cord blood
- Serum, plasma, buffy coat
- Peripheral blood mononuclear cells (PBMC)
- Samples for Microbiome Research (stool, skin, etc.)
- Urine
- Stool
- Saliva
- DNA and RNA
- FFPE tissue
- Fresh tissue
- Frozen tissue
- TMA (tissue microarray) library
- Histology facility
- General histology
- Immunostaining
- TMA (tissue microarray) construction and staining
- Digital scanning and image scoring
- Clinical and epidemiologic questionnaire data associated with banked samples
Services
Please contact BLSAdmin@RoswellPark.org for a narrative and project-specific cost estimate.
We procure, process and store peripheral blood, bone marrow and other biological samples from patients with a variety of solid and hematological malignancies for researchers at Roswell Park and other institutions. We strive to provide specimens that are rigorously collected, processed, and stored so that the integrity of potential analytes will be consistent across all patients and controls. Samples are drawn and sent to the BLS laboratory for processing into serum, plasma, buffy coat, peripheral blood mononuclear cells (PBMC), bone marrow mononuclear cells (BMMCs) and genomic DNA.
Genomic DNA is readily available from extracted blood using an Autogen XTRACT 16+ nucleic extractor which allows rapid extraction from up to 48 samples per day, a Qubit fluorometer from ThermoFisher that facilitates accurate quantification and evaluation of double- stranded DNA.
Samples are processed following stringent standard operating procedures and stored in barcoded containers that are linked and managed in our Laboratory Information Management System (LIMS). The liquid bank has more than 1.3 million blood and/or other sample types banked from more than 43,000 patients and healthy controls. This includes more than 220,000 cryopreserved marrow and blood samples from patients with hematological malignancies dating back to 1991. Banking of blood specimens from patients with solid tumors started in 2003. All samples have related clinical data available.
Tissue
Biospecimens are available for fresh, frozen, and for Formalin Fixed Paraffin Embedded (FFPE) tissue collected by the tissue procurement lab. Tissue microarrays (TMAs) are built using the remnant blocks from the Department of Pathology archive resource (PAR). All tissue used for research is collected under remnant tissue protocol I 115707. Currently, tissue samples are requested and sent to Genomics Shared Resource to extract DNA or RNA.
DNA and RNA
DNA and RNA are isolated from tumor and matching non-tumor tissues in the biobank. DNA and RNA are primarily extracted from frozen tissue samples, but DNA can be extracted from FFPE tissue as well. All samples are QC’ed after extraction to confirm the DNA is of high quality.
FFPE tissue
Formalin-fixed paraffin-embedded (FFPE) blocks from surgical pathology tissues are archived and can be accessed for research purposes. Our archive of paraffin tissues is extensive, having approximately more than 200,000 cases dating back to 1993. This database can be electronically searched and used for research purposes.
Fresh and frozen tissue
Fresh and frozen remnant tissues from surgical specimens may be distributed for research purposes. Our biospecimens are procured by certified pathology assistants and then stored in -80C freezers that are protected by state-of-the-art monitoring systems and 24/7 emergency contact schedules.
Requests for fresh or frozen tissues are guided by the Laboratory Information Management System (LIMS) and are integrated with other shared resource facilities (e.g., the Genomics Shared Resource).
The number of biospecimens available in the biobank varies by disease site and their utilization. Specific details for samples in the biobank including inventory, surgical events, tumor dimensions, and the numbers of tissues procured, banked, and distributed per disease site are available on the LIMS dashboard. External users, please contact BLSAdmin@RoswellPark.org for further information.
TMA library
Tissue microarrays (TMAs) are constructed by using a hollow needle to remove very small tissue cores from multiple tumors of interest which are then inserted into a recipient paraffin block in an arrayed fashion. This format permits the screening of a large number of patient samples on a single slide. TMAs are an efficient and effective way to screen potential biomarkers and are typically used in conjunction with immunohistochemical (IHC) staining or fluorescent in situ hybridization.
The BLS has constructed more than 200 different blocks representing a wide variety of disease and tissue types from more than 16,000 patients.
The Histology Facility is a centralized lab that provides services for standard tissue processing and embedding, cryotomy, microtomy, general histology staining, antibody optimization and immunohistochemistry (IHC) for both human and animal tissues. Histology staff work closely with the Biomedical Research Informatics Shared Resource (BRISR) and other shared resource labs to identify samples sets, and then perform the required staining for research studies that use IHC. The lab uses several auto stainers for consistent and high-throughput IHC staining. Antibody staining procedures and IHC slides are regularly reviewed by a pathologist for quality control. The histology facility continually expands the repertoire of antibody protocols, and have over 500 procedures.
The following histology services are provided by the Histology Facility:
- Immunohistochemical staining
- Antibody optimization for immunohistochemistry
- Hematoxylin and eosin staining
- Special histological staining (e.g., GMS, Masson’s Trichrome, PAS)
- Cryotomy
- Microtomy
- Paraffin embedding
- Standard tissue processing
- Custom TMA construction (human or pre-clinical model samples)
- Tissue prep for Visium (collaboration with Genomics Shared Resource)
- Tisue prep for Phenocycler (collaboration with Flow and Immune Analysis Shared Resource)
- Sterile needle coring for DNA/RNA isolation
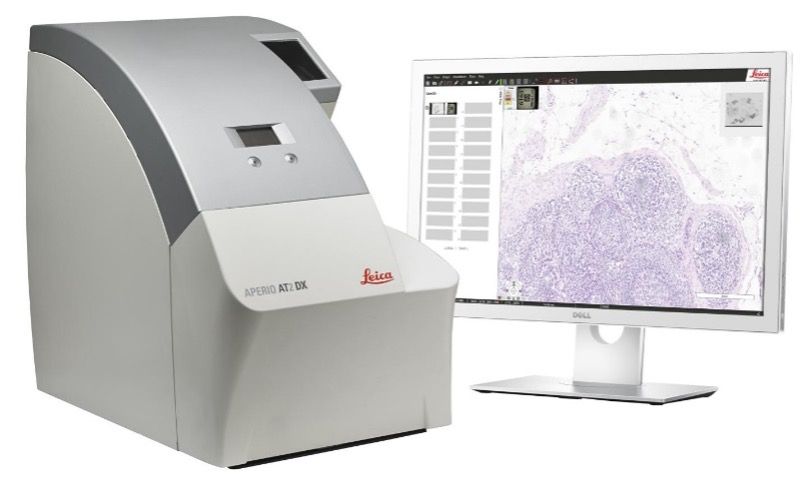
The Imaging and Analysis lab uses an Aperio® AT2 slide scanner. Slide scanning permits investigators to easily access their digital images of slides from anywhere using Aperio® eSlide Manager, a web-based digital pathology information management system. Annotation of the digital slides can be conducted via a digital platform using Aperio® ImageScope. This allows researchers to create digital images for publications and facilitates inter-institutional collaborations by providing rapid and easy sharing of digital image information.
AT2 system
The AT2 is an ultra-fast, high-capacity scanning system with powerful 400-slide capacity. This system creates digital images from glass slides with superior image quality and includes the following features:
- 20X and 40X scanning magnification capabilities
- Create digital slides in multiple formats (SVS, JPEG, TIFF, composite web slide)
- View and edit digital slides with the user-friendly and freely downloadable ImageScope viewing software
- Connect multiple, remote parties through digital slide conferences.
ImageScope software
ImageScope is a freely downloadable viewing software for digital slide viewing. ImageScope is easy to use and puts a host of powerful capabilities at your fingertips.
- View digital slides from any workstation on the network; this eliminates the need to physically transport glass slides instantly pan and zoom to any region of the slide.
- Built-in web conferencing: real-time digital slide sharing and discussion in multiple remote locations.
- View multiple digital slides concurrently.
- Apply image adjustments in real time for contrast, brightness and gamma.
- Analyze entire digital slides or selected regions.
- Add graphical and text annotations to digital slides.
- Use the built-in ruler to make on the spot measurements.
- Save a region or selected regions of a digital slide to a file.
- Allows researchers to create digital images for publication as well as facilitate inter-institutional collaboration by providing rapid and easy sharing of digital image information.
- ImageScope also enables side-by-side coordinated viewing of multiple slides, something that cannot be done with traditional glass slides. In addition, Aperio's SmartSync™ feature makes it easy to compare different sections from the same tissue block, even when sections are rotated or translated with respect to one another.
Leica eSlide Manager (eSlide)
eSlide is a web-based digital pathology imaging and information management system. Access to eSlide can be granted by completing an application.If you would like to request access to eSlide, contact BLSImaging@RoswellPark.org.
Automated image analysis
Image analysis is a service also provided by the BLS. Algorithms can be tailored to fine-tune the cellular, nuclear, and stain parameters, creating an optimized algorithm macro for each antibody target and tissue type to select the cells of interest. The original digital slide image is never modified. Rather, an annotation layer with the markup image and quantitative data is created and linked to the digital image. Currently, we have one full-time employee with expertise in digital slide analysis. The BLS performs image analysis for slides stained by IHC only. Nuclear, cytoplasmic and membranous expression can be scored individually or in any combination. Vascular analysis can be performed and tailored to calculate specific vessels by tweaking vessel size parameters. Some dual stains can also be analyzed with this system depending on the type of stains and target.
Epidemiological data
At the time of consent, participants (patient or non-patient/control) are asked to complete a self-administered epidemiological questionnaire available online or paper version. The questionnaire contains more than 1,100 items including:
- Demographics and background
- Personal and family history of cancer and other conditions
- Medical and cancer screening histories
- Lifestyle (smoking, diet, physical activity, alcohol, cannabis)
- Supplement and medication use
The questionnaire data dictionary and codebooks are available to investigators for research planning and analysis.
Clinical data
Each sample collected from patients has clinical data available. The clinical data is available from disease specific data managers, BRISR, and BLS data managers. Available data includes:
- Roswell Park Cancer Registry data with basic tumor information, treatment and first recurrence details. Data collected includes: diagnosis date, laterality, histology, behavior, grade, tumor size, clinical and pathological stage, distant sites, tumor markers, surgery date, pathology and recurrence (as of the sample collection date).
- Electronic health record: Includes drug treatment, patient information (height, weight, BMI, blood pressure, etc.), visits, orders (scans, scopes, tests, procedures, etc.), comorbidities, family history, karyotype, mutational profile, and outcome.
- Additional medical record abstraction
- Honest broker services
- Linking and de-identifying data and study data file storage
Recruitment and procurement
BLS has a team devoted to participant recruitment, located within Registration in the main hospital lobby in downtown Buffalo and throughout hospital clinics. Their focus is to perform informed consent on patients and healthy controls. The Universal consent includes questions for approval for biospecimen procurement including blood collections, bone marrow and remnant tissue along with access to clinical data, and permission to recontact for additional research opportunities and/or additional collections. Once consented, recruitment staff can order blood samples and track for serial samples to be collected alongside existing clinical laboratory orders and drawn by the phlebotomy service at Roswell Park. Bone marrow and tissue collections are part of routine clinical collections and surgery.
Family members and friends accompanying patients, as well as other visitors to Roswell Park, also are asked to donate samples as controls. Control samples follow the same sample procurement and processing procedures as patient samples.
Roswell Park Staff may reach BLS Recruitment Coordinators for recruitment issues by paging “Biorepository Consent" from inside the hospital. Recruitment staff may also be reached for questionnaire issues by telephone at 716-845-7774 or please email BLSRecruitment@RoswellPark.org.
Frequently asked questions
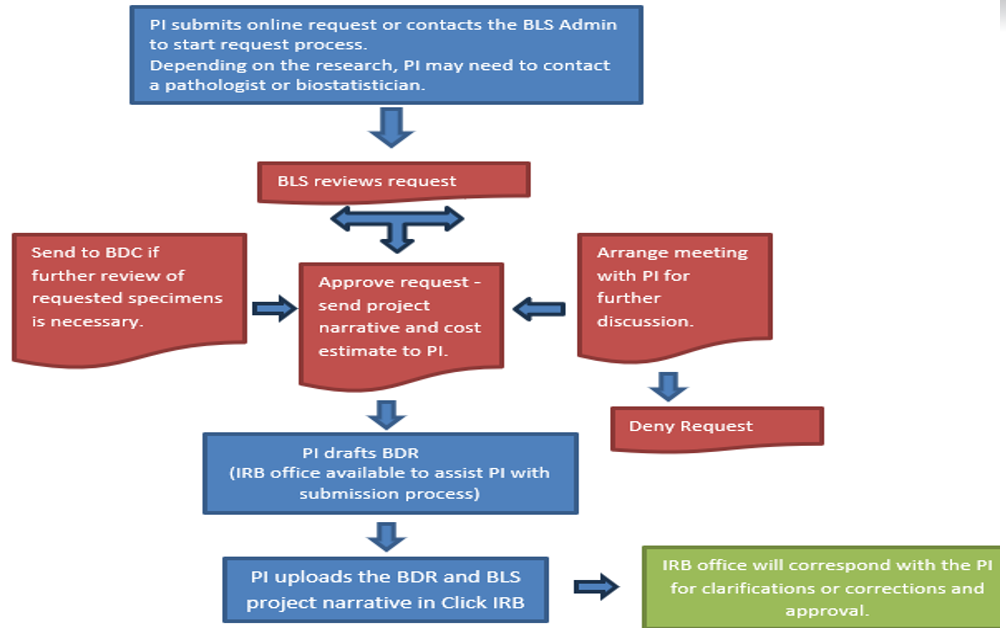
Generally, every time biospecimens are being used for analysis with the purpose of exploring a hypothesis and publishing findings an IRB approval is needed. This could be done either as a Biospecimen and Data Research (BDR) project or non-interventional, investigator initiated clinical trial. Please submit a request to start the request process.
Please submit a request to start the request process. Once a request is submitted, you will be contacted for follow-up. Following the review of your request, BLS will provide a project narrative to include with your BDR application, a copy of the application, a cost estimate, and a list of the next steps to get your BDR approved as outlined in the chart. While BLS works closely with the TRGs, TRG approval is not necessary for a biospecimen request. Biospecimen and Data Committee will review biospecimen requests as needed for utilization.
Please submit a request to start the request process. If you are looking to get basic information about available biospecimens for a cohort of patients, we recommend utilizing nSight.
Please contact BLSImaging@RoswellPark.org.
Yes. Please include the following statement in the acknowledgements section of your manuscript: This work was supported by grants from the National Cancer Institute (NCI) grant P30CA016056 involving the use of Roswell Park Comprehensive Cancer Center’s Biorepository and Laboratory Services Shared Resource.
Sample and Data Management
Amy Betz
Stephanie Farnham
Ashley Wilson
BLS Laboratory
Patricia Brown
Casey Flatt
John Hamilton, PhD
Alyse Kalota, MS
Nancy Kelly
Nicholas Kisiel
Hannah Kunes
Linda Lutgen
Nikkita Maybach
Leonard Medico Jr.
Steven Minderler
Joseph Moberg
Amrutha Mohan, MS
Priya Nair
Kelsey Paine
Carol Siegel, MA, CCRP
Leighton Stein
Brady Thompson, MA
Erika VanDette
Haley Ward, MA
Cassandra Whalen
Recruitment
Email: BLSRecruitment@RoswellPark.org
Deisy Moran
Jaina Ruffino
Thomasina Smith
Erin Striegel
Location & hours
Roswell Park Comprehensive Cancer Center
Biorepository & Laboratory Services (BLS) Shared Resource
Gratwick Basic Science Building, 6th & 7th Floors
Elm and Carlton Streets
Buffalo, New York 14263
BLS Recruitment Service: Monday – Friday, 7 a.m. – 5:30 p.m.
Laboratory Service: Monday – Friday, 6 a.m. – 6 p.m.
Data Service: Monday – Friday, 7 a.m. – 5 p.m.
This shared resource is funded by NCI P30CA16056 and NIH C06OD037775. Publications should cite the core grant in the acknowledgment section, if publications use samples and data provided by the shared resource.
An example: Data, samples, and Honest Broker services for this study were provided by the Biorepository and Laboratory Services (BLS) which is funded by the National Cancer Institute (NCI P30CA16056) and ONE Biorepository Grant (NIH C06OD037775) and is a Roswell Park Comprehensive Cancer Center Support Grant shared resource.
One Biorepository and Laboratory Services (BLS) is a comprehensive data and biospecimens bank (tumor tissue and their derivatives, blood, and bone marrow) for research along with a range of services that accompany biospecimens. Functionally the BLS is divided into biospecimen procurement and banking, histology services, digital scanning, and image scoring.
The biospecimen procurement and banking section of BLS consists of procured tissue, blood, and bone marrow samples. The tissue subsection of the BLS was presented at the annual meetings of the American Association of Cancer Institutes (AACI) as a model of excellence. All biospecimen tissue procurement is performed by trained, certified, Pathologist Assistants using detailed standard operating procedures (SOPs).
The blood and bone marrow biorepository section offers access to cryogenically preserved plasma, serum, buffy coat, PBMC, DNA, whole blood, cord blood, and bone marrow. The blood samples are linked to epidemiologic questionnaire data and clinical information for investigators conducting translational research related to cancer prevention, etiology, detection, and treatment. The BLS follows QC guidelines outlined in Best Practices for Repositories by ISBER (International Society for Biological and Environmental Repositories) and NCI Best Practice Guidelines as well as all Roswell Park biosafety policies and procedures. Quality assurance (QA) guidelines cover the systematic monitoring and evaluation of all aspects of workflow processes. With the Universal Consent, BLS also assists in conducting studies with targeted patient populations at Roswell Park and provides infrastructure for banking samples from extramurally funded studies.
Equally important, the histology subsection provides general histology, immunostaining, and other specialized tissue staining services. Accompanying digital scanning and image scoring services are also available, jointly overseen by a pathologist and trained imaging specialist with more than 10 years of experience in the field. The resource also has extensive experience with handling fresh, frozen, or formalin-fixed paraffin-embedded tissues from human, mouse, or rat sources. Immunohistochemistry is a major component of the BLS with over 50 new antibody protocols optimized annually and over 900 antibody staining procedures available for research requests. Other services available include clinical block archiving, review and retrieval services, tissue microarray construction, Aperio slide scanning, and laser microdissection services. BLS plays an essential role at Roswell Park helping investigators establishing Biospecimen and Data Research (BDR) protocol and supports the submission of grants by providing feasibility analysis, cost estimates and letters of support.
All banking activities and requests are managed through the electronic Laboratory Information Management System (LIMS). The LIMS documents procedures including record control, handling and storage, internal auditing, and overall improvement measures. All instruments, specimens, and data are connected to a secure Roswell Park server where all data may be stored long term and accessed anywhere with the proper credentials. Biospecimens are stored in -80°C and liquid nitrogen freezers that are protected by state-of-the-art monitoring systems with backup. This shared resource facility has 33 staff members and occupies more than 10,600 sq. ft. of research-dedicated space.
As an early contributor to The Cancer Genome Atlas (TCGA), Roswell Park was a major Tissue Source Site (TSS) also providing matched genomic DNA. Roswell Park was the 6th leading contributor of specimens with linked clinical data to the TCGA. BLS also made substantive contributions to similar research efforts, providing matched tissue and blood along with associated data. This includes but not limited to, the National Mesothelioma Virtual Bank (NMVB), African American Breast Cancer Epidemiology and Risk (AMBER) Consortium, Oncology Research Information Exchange Network (ORIEN), and NCI Patient-Derived Models Repository (PDMR).
Cancer Center Support Grant (CCSG) Program members with peer-reviewed funding are given top priority for requests, followed by Roswell Park investigators with newly developing programs, then external academic investigators with federal funding and finally commercial investigators. Fees for cost recovery are tiered for these different groups of investigators.