Next-generation sequencing (NGS) has become widely accessible in research and clinical laboratories. By using whole-genome, whole-exome and transcriptomes sequencing, we can detect various types of variants such as single-nucleotide variants (SNVs), small insertions/deletions (Indels), as well as large variants including structural variants (SVs) and copy-number variants (CNVs).
Integrative analysis of all types of variants can reveal frequently mutated genes and pathways in specific cancer types, as well as potential association with clinical phenotypes and treatment outcomes. These findings provide new insights into the mechanisms of various types of cancers, and highlight potential biomarkers for therapeutic development.
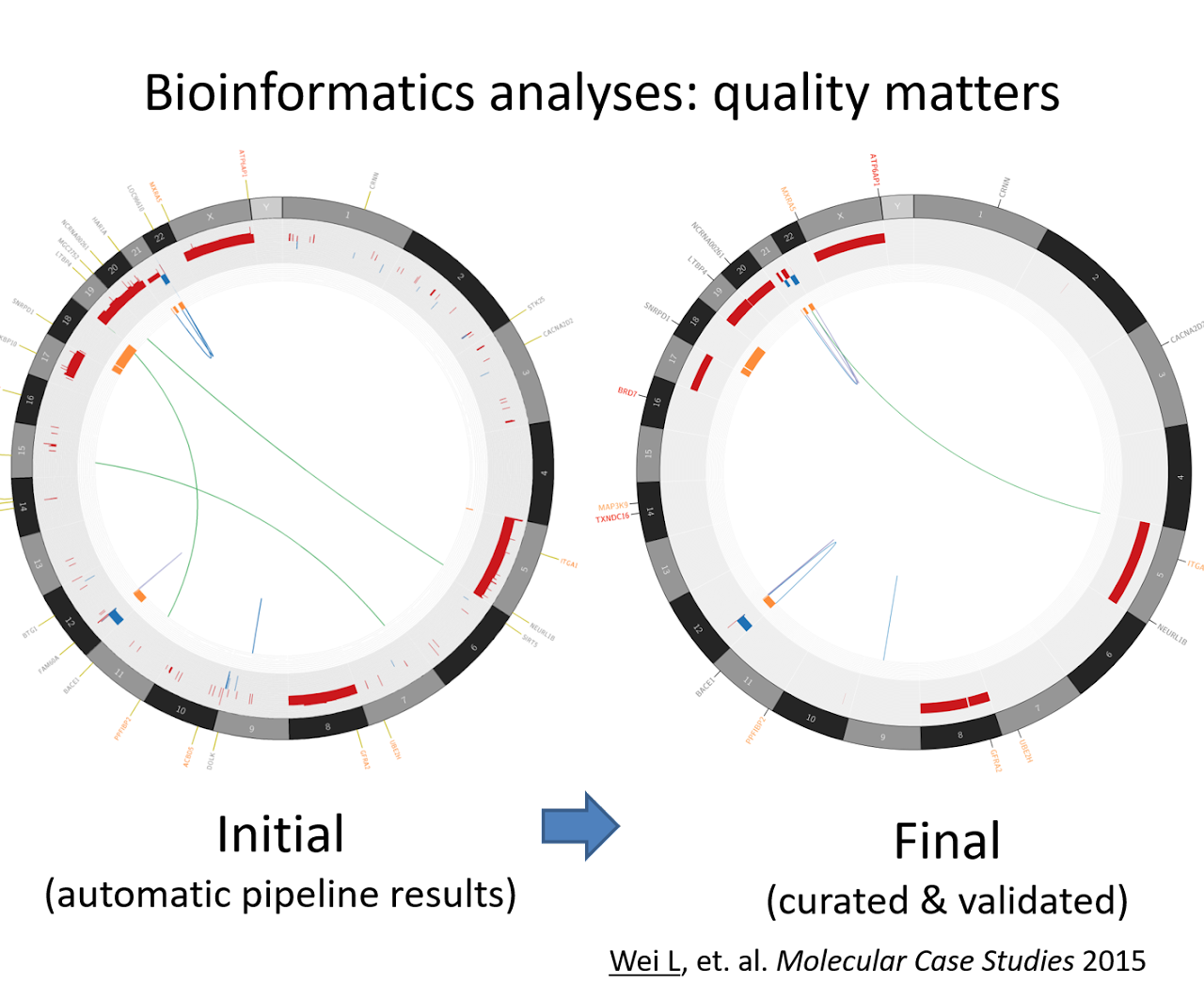
With many tools available, accuracy is the key to high-quality data analysis. Due to various types of potential issues in the process of sampling/sequencing/mapping, it is not uncommon to see both false positive and false negative generated in automatic predictions. After fixing these errors, the mutational profile may look dramatically different.
Selected papers
Wei L, Wang J, Lampert E, Schlanger S, DePriest AD, Hu Q, Gomez EC, Murakam M, Glenn ST, Conroy J, Morrison C, Azabdaftari G, Mohler JL, Liu S, Heemers HV. Intratumoral and Intertumoral Genomic Heterogeneity of Multifocal Localized Prostate Cancer Impacts Molecular Classifications and Genomic Prognosticators. Eur Urol. 2017 Feb;71(2):183-192. doi: 10.1016/j.eururo.2016.07.008. Epub 2016 Jul 21. PMID: 27451135; PMCID: PMC5906059. - Editorial highlights
Wei L, Liu S, Conroy J, Wang J, Papanicolau-Sengos A, Glenn ST, Murakami M, Liu L, Hu Q, Conroy J, Miles KM, Nowak DE, Liu B, Qin M, Bshara W, Omilian AR, Head K, Bianchi M, Burgher B, Darlak C, Kane J, Merzianu M, Cheney R, Fabiano A, Salerno K, Talati C, Khushalani NI, Trump DL, Johnson CS, Morrison CD. Whole-genome sequencing of a malignant granular cell tumor with metabolic response to pazopanib. Cold Spring Harb Mol Case Stud. 2015 Oct;1(1):a000380. doi: 10.1101/mcs.a000380. PMID: 27148567; PMCID: PMC4850888.
Holmfeldt L, Wei L, Diaz-Flores E, Walsh M, Zhang J, Ding L, Payne-Turner D, Churchman M, Andersson A, Chen SC, McCastlain K, Becksfort J, Ma J, Wu G, Patel SN, Heatley SL, Phillips LA, Song G, Easton J, Parker M, Chen X, Rusch M, Boggs K, Vadodaria B, Hedlund E, Drenberg C, Baker S, Pei D, Cheng C, Huether R, Lu C, Fulton RS, Fulton LL, Tabib Y, Dooling DJ, Ochoa K, Minden M, Lewis ID, To LB, Marlton P, Roberts AW, Raca G, Stock W, Neale G, Drexler HG, Dickins RA, Ellison DW, Shurtleff SA, Pui CH, Ribeiro RC, Devidas M, Carroll AJ, Heerema NA, Wood B, Borowitz MJ, Gastier-Foster JM, Raimondi SC, Mardis ER, Wilson RK, Downing JR, Hunger SP, Loh ML, Mullighan CG. The genomic landscape of hypodiploid acute lymphoblastic leukemia. Nat Genet. 2013 Mar;45(3):242-52. doi: 10.1038/ng.2532. Epub 2013 Jan 20. PMID: 23334668; PMCID: PMC3919793. - Cover story