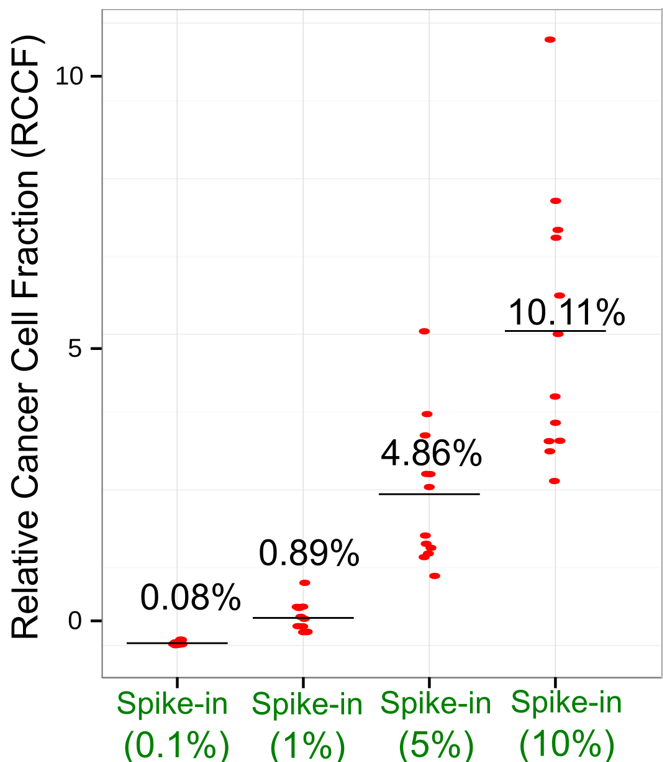
We’ve conducted innovative studies aiming to expand our detection limits to very-low-frequency (<5%) mutations, which are important for cancer early detection and residual disease monitoring.
The technical challenge comes from distinguishing real somatic mutations from background noise caused by sequencing, mapping, or library preparation errors. We were able to overcome this challenge by using bioinformatics algorithms with in-silico error suppression.
The accuracy of our detection method was validated using spike-in experiments.
Collaborations
Collaboration with Roswell Park’s Dr. Gyorgy Paragh, Dr. Wendy Huss, and Dr. Barbara Foster, and Dr. Sean Christensen of Yale University.
Most cancers are caused by a lifetime accumulation of somatic mutations. Despite the important roles they play in tumor initiation and progression, mutations in physiologically normal tissues have been under-studied due to their low abundance and random patterns.
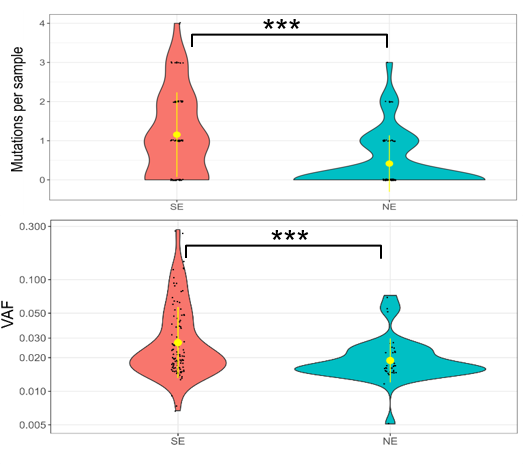
We sequenced a large number (n = 450) of normal skin samples to identify early clonal mutations (CMs). The skin samples were collected in an individual-matched manner containing equal numbers of sun-exposed and non-sun-exposed samples from every donor, which allowed us to exclude mutations caused by other factors such as aging.
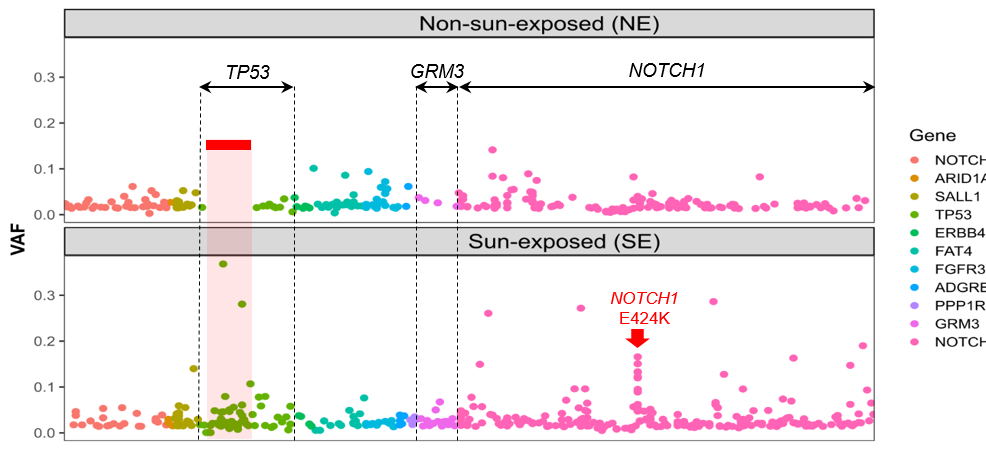
This allowed us to then precisely identify the mutation patterns introduced by UV light. The results not only confirmed the previous findings of widespread mutations in aged skin, as well as elevated mutational burden after UV-exposure, but also identified several associated novel mutational patterns.
For instance, we found UV-induced mutations were enriched in specific genomic regions, including several mutational hotspots where mutations are highly recurrent only in UV-exposed skin samples.
These findings pave the road for future development of a quantitative measurement of UV-induced DNA damage, which allows objective assessment of skin cancer risk as well as early intervention such as field treatment.
Wei L, Christensen SR, Fitzgerald ME, Graham J, Hutson ND, Zhang C, Huang Z, Hu Q, Zhan F, Xie J, Zhang J, Liu S, Remenyik E, Gellen E, Colegio OR, Bax M, Xu J, Lin H, Huss WJ, Foster BA, Paragh G. Ultradeep sequencing differentiates patterns of skin clonal mutations associated with sun-exposure status and skin cancer burden. Sci Adv. 2021 Jan 1;7(1):eabd7703. doi: 10.1126/sciadv.abd7703. PMID: 33523857; PMCID: PMC7775785.
Collaboration with Roswell Park urologist Dr. Khurshid Guru.
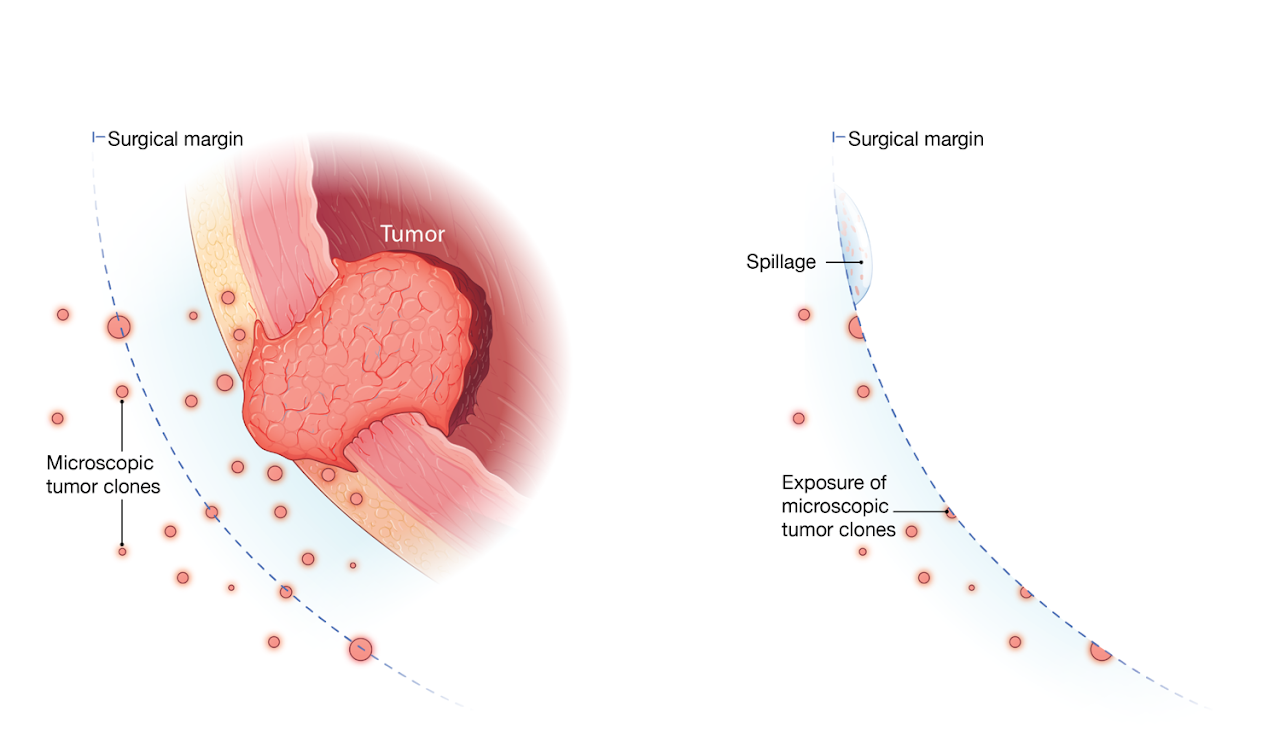
Bladder cancer is a common cancer type with high recurrence rate after surgery, while the mechanism of recurrence remains unclear. One major hypothesis is that the tumor spillage during surgery may result in residual tumor cells (RTCs), which may be responsible for tumor recurrence.
However, RTCs could not be detected in previous tests using conventional mRNA markers or cytology. In collaboration with the surgical urology team, we designed a comprehensive study that collected pelvic washings at multiple time points during the surgery, followed by ultra-deep sequencing to measure RTCs in each washing.
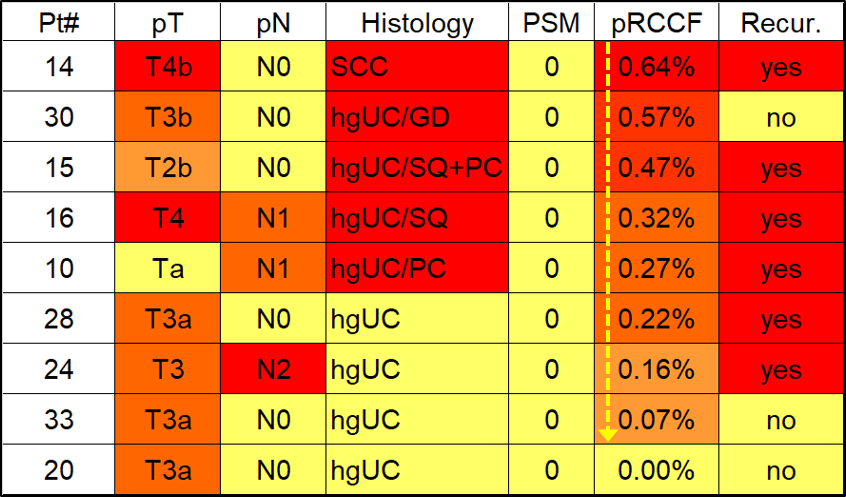
The results were remarkable: all washings before the surgery were negative for RTCs; however, most patients’ post-surgery washings tested positive, confirming the presence of RTCs after surgery.
Importantly, we found a significant association between the levels of RTCs and the aggressiveness of the tumor’s histology, suggesting the identified RTCs were unlikely to be caused by random tumor spillage during surgery, but were probably related to preexisting infiltrating microscopic tumor clones.
In the current data, we found that the levels of RTCs were more significantly associated with cancer recurrence than conventional markers, such as tumor grade or surgical margin, which may open a new area of developing novel, highly sensitive biomarkers to predict recurrence.
Wei L, Hussein AA, Ma Y, Azabdaftari G, Ahmed Y, Wong LP, Hu Q, Luo W, Cranwell VN, Bunch BL, Kozlowski JD, Singh PK, Glenn ST, Smith G, Johnson CS, Liu S, Guru KA. Accurate Quantification of Residual Cancer Cells in Pelvic Washing Reveals Association with Cancer Recurrence Following Robot-Assisted Radical Cystectomy. J Urol. 2019 Jun;201(6):1105-1114. doi: 10.1097/JU.0000000000000142. PMID: 30730413; PMCID: PMC6784327.
Collaboration with Roswell Park pathologist Dr. Carl Morrison.
The normal tissue adjacent to tumor tissue is often used as the normal DNA source in somatic mutation detection for filtering out germline polymorphisms. One problem is these normal tissues frequently contain low-level tumor cells, which would lead to false-negative calls as somatic mutations can be misclassified as germline SNVs when also present in normal tissue.
By using ultra-deep sequencing, we confirmed the presence of low-level tumor contamination in normal tissue, and found the observed contamination was related to inadvertent handling during the surgical pathology gross assessment and tissue procurement process.
This finding highlights the acquisition of high-quality normal tissue as one important but often overlooked area in somatic mutation detection.
Wei L, Papanicolau-Sengos A, Liu S, Wang J, Conroy JM, Glenn ST, Brese E, Hu Q, Miles KM, Burgher B, Qin M, Head K, Omilian AR, Bshara W, Krolewski J, Trump DL, Johnson CS, Morrison CD. Pitfalls of improperly procured adjacent non-neoplastic tissue for somatic mutation analysis using next-generation sequencing. BMC Med Genomics. 2016 Oct 19;9(1):64. doi: 10.1186/s12920-016-0226-1. PMID: 27756300; PMCID: PMC5070097.
Dr. Wei also has an ongoing collaboration with Roswell Park’s Dr. Philip McCarthy and Dr. Jens Hillengass, and the University of Tennessee’s Dr. Swapna Thota to characterize Clonal hematopoiesis of indeterminate potential (CHIP) identify biomarkers for secondary leukemia and other blood disorders.
![nadvertent handling during sample grossing and tissue procurement results in tumor contamination in the adjacent normal tissue]](/sites/default/files/styles/max_1300x1300/public/2022-03/Research2-Figure6.png?itok=kT-JphWc)