Widely recognized that binding between proteins is central to virtually all biological processes. With several completed genome sequences as a framework with which to interpret such interactions, several large scale projects have attempted to define protein interactions for all of the open reading frames (ORFs) of simple organisms including viruses, bacteria, yeast, Drosophila, and C. elegans.
Although other methods of defining protein interactions are possible, the most highly developed method for genome-wide analysis is the original yeast two-hybrid system in which interactions are monitored by the induction of gene expression.
Large scale projects to define all of the interactions occurring between all of the ~6,000 ORFs in yeast have been accomplished using the yeast two hybrid system. However, application of this technology to mammalian genomes, which contain on the order of 10-fold greater complexity, is currently not feasible due to the exponentially greater number of potential interactions that must be scored. Thus, there is a need for an efficient method of identifying genome-wide protein interactions for organisms with complex protein interactions.
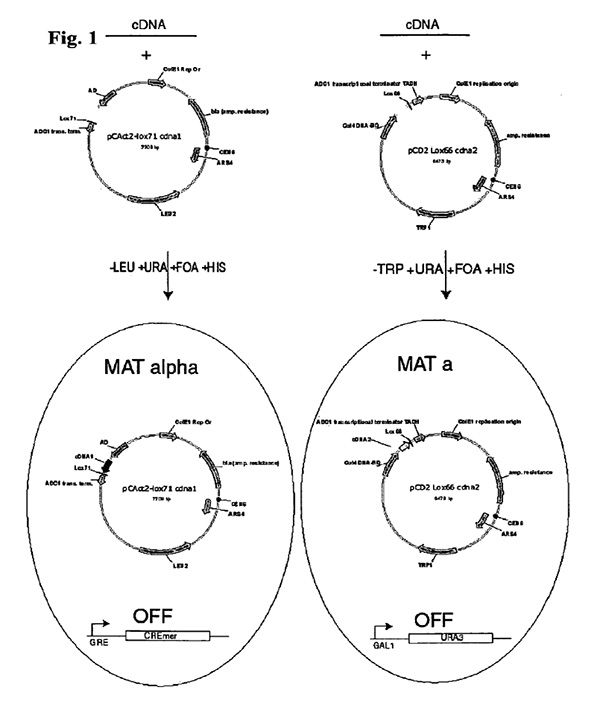
The present invention provides a modification of two-hybrid technology that permits the identification of many pairs of interacting proteins. The interacting proteins comprises first and second test proteins with interact with each other in a cell. The invention provides a plasmid pair for use in a modified two hybrid system where the first plasmid comprises a coding sequence for a DNA binding domain (DBD) of a transcription activator and the second plasmid comprises a coding sequence for a transcription activation domain of a transcription activator (AD), and each plasmid further comprises a recombinase recognition site.
This invention also provides a vector system and a method for establishing a comprehensive protein interaction map from a cDNA library by adapting two hybrid technologies to allow physical linkage of cDNAs encoding interacting proteins and to improve the efficiency of identifying interacting cDNA sequences by modifications allowing the application of a modified serial analysis of gene expression (MAGE).