Leveraging multi-omics approaches to study determinants of lineage plasticity in cancer
Under chronic stress or therapeutic pressure, cells undergo epigenetic reprogramming away from defined lineage states towards more primitive states with enhanced pluripotent potential. Identifying mechanisms that drive this lineage plasticity is vital in understanding how tumors develop and evade therapeutic response.
Using omics scale data to investigate and quantify plasticity
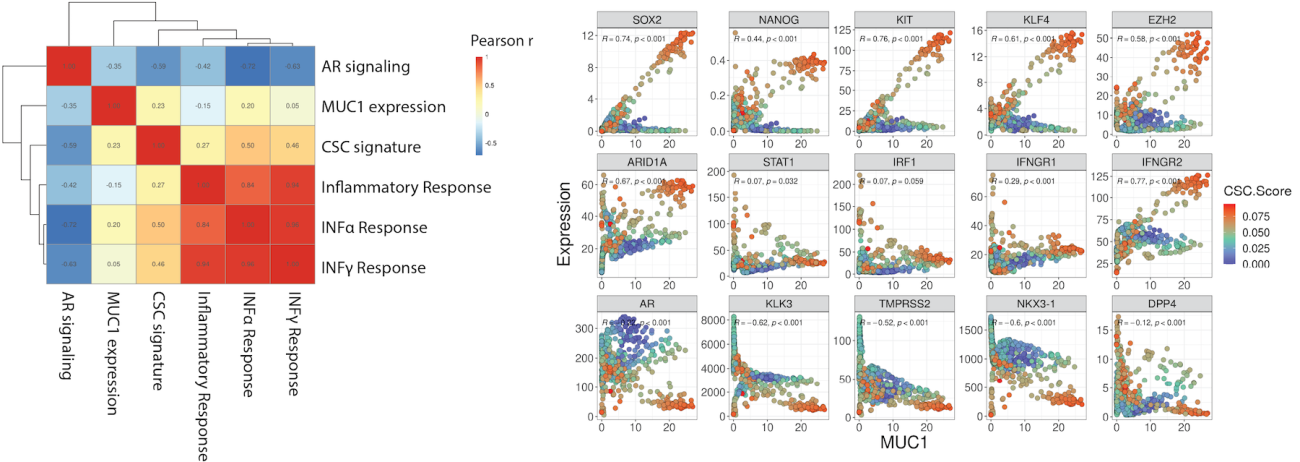
Integrative analysis (RNA-seq, ATAC-seq) of preclinical models and publicly available bulk and single-cell human tumor data has revealed roles of MUC1 in driving stemness and intracellular interferon signaling in various tumor types, including castration-resistant prostate cancer (Figure 1) and non-small cell lung cancer.
- Hagiwara M, et al. MUC1-C integrates type II interferon and chromatin remodeling pathways in immunosuppression of prostate cancer. Oncoimmunology. 2022;11(1):2029298. (Co-corresponding author)
- Fushimi A, et al. Dependence on the MUC1-C Oncoprotein in Classic, Variant, and Non-neuroendocrine Small Cell Lung Cancer. Mol Cancer Res. 2022 Sep 2;20(9):1379-1390. doi: 10.1158/1541-7786.MCR-22-0165. (Co-corresponding author)
Experimental and computational analysis determined a role of NCOR2 in suppressing response to androgen deprivation therapy in prostate cancer models. Integrative multi-omics revealed interactions of NCOR2 with pioneering transcription factors that had important roles in governing global transcription, DNA methylation and super-enhancer function.
Further, we leveraged whole-genome bisulfite sequencing to identify regions of dynamic methylation in the normal prostate epithelial differentiation program, which we subsequently revealed were aberrantly methylated in prostate cancer.
- Long MD, et al. Reduced NCOR2 expression accelerates androgen deprivation therapy failure in prostate cancer. Cell Rep. 2021 Dec 14;37(11):110109.
- Long MD, et al. Dynamic patterns of DNA methylation in the normal prostate epithelial differentiation program are targets of aberrant methylation in prostate cancer. Sci Rep. 2021 Jun 1;11(1):11405.
Connect with the Long Lab
Email: Mark.Long@RoswellPark.org
Phone: 716-845-2541
Department of Biostatistics and Bioinformatics
Roswell Park Comprehensive Cancer Center
Elm and Carlton Streets
Buffalo, NY 14263