What we do
The Research Information Technology Department is comprised of a dedicated group of professionals focused on meeting the technology needs of the research community at Roswell Park Comprehensive Cancer Center. Our mission is to understand and apply current and cutting-edge technologies to facilitate, enhance and accelerate oncology research. The software tools, applications, platforms, technologies and services provided support a wide range of clinical, basic science, translational and population health research.
Communication, collaboration and partnership with investigators and research support departments are paramount. Much effort is focused on ensuring these happen effectively so that the appropriate technologies can be leveraged and applied. Included in these technologies and services provided are hardware, software, storage, high-performance computing, application support, training, data sharing and data analytic services.
Technology
Infrastructure
The Research IT Department ensures the latest information technologies are made available to the research community. The backbone infrastructure layer of the organization consists of Cisco gigabit ethernet, 1,300+ servers, 5,500 workstations (Windows/Mac OS/Linux) and ~4 PB total storage.
High-Performance Computing
For computationally intensive work, researchers have access to the High-Performance-Computing (HPC) resources located at the University at Buffalo Center for Computational Research (CCR). These resources consist of more than 30,000 processing cores and 4.7 PB of high-performance storage. The CCR is a 24/7 state-of-the-art high-performance computing center and is a leading academic supercomputing facility with support staff that have expertise in computing, visualization and networking. There are 15 dedicated compute nodes that are integrated into the CCR compute environment and are dedicated to Roswell Park researchers. This includes GPU nodes to support artificial intelligence and machine learning applications, and a total of 1 PB of data storage.
Software applications
We provide direct support for a wide variety of software applications designed to meet the business and scientific needs of the researchers, shared resources and other research supporting departments. The suite of software products are vendor-based or in-house developed applications that scale from small off-the-shelf tools to large institution-level applications.
Types of services provided
Our primary focus is to provide a wide range of technologies and technology services that support, facilitate, and enhance the research activities of the organization, research departments and individual researchers. Below is a summary of the services and technologies provided:
The Research IT Department provides the software tools, technologies and services required to support the entire clinical research lifecycle. This is accomplished by procuring, implementing, enhancing and supporting enterprise-level, vendor-based software products. The suite of tools provided meet the specific needs and requirements of the investigators and the clinical trial support departments. Where required by regulation, the tools undergo an FDA validation process to ensure they adhere to the FDA requirements for maintaining electronic records and signatures (FDA 21 CFR Part 11).We collaborate closely with the Office of Research Protection and Clinical Research Services to continually support and enhance the tools and technology framework required for clinical research activities.
We provide software tools, technologies and services to support a wide range of biospecimen and data research. Both vendor-based and custom in-house developed software applications have been implemented to meet the needs of the individual investigator, research laboratory and shared resource. We are fully support a large enterprise-level Laboratory Information System (LIMS). Services include providing analysis, design, development, testing and implementation of the LIMS software to meet the unique workflows and data management needs of the laboratory or shared resource. For cohort building and data discovery, the team has developed and fully supports custom software applications to enhance and facilitate these activities (i.e., nSight, REVEAL, Imagine).
Through a formal collaboration with the Center for Computational Research (CCR) at the State University of New York at Buffalo (UB), our researchers have access to the high-performance computing cluster (HPCC) with more than 8,000 processor cores and accompanying high-performance storage. The CCR is a leading academic supercomputing facility, maintaining a high-performance computing environment, high-end visualization laboratories and support staff with expertise in computing, visualization and networking.
We provide a host of services to assist researchers in utilizing these HPCC resources. These services include: onboarding new users, introductory training in scientific programming and machine learning, file transfer and data management, software package licensing, installation and maintenance.
The department provides custom in-house developed tools and technologies to the research community to facilitate data mining and research cohort discovery. The nSight software tool is a web-based application that allows researchers to explore and analyze data from multiple clinical and research data sources. This allows users to develop research cohorts for research ideas and related grants, study design, publications, etc. The nSight tool allows the users to identify and save study cohorts based on patient, tumor, treatment and other clinical or research characteristics. The tool rapidly accelerates these overall processes.
We provide services and technologies to assist researchers with performing digital image analysis research. Custom, in-house developed tools, along with image analysis pipelines, provide a framework for initiating and executing this type of research. We offer computational science expertise to help assess and guide investigators with the initiation and execution of this type of research. The tools and pipelines offered allow researchers to efficiently build study cohorts, search PACS archive, download and deidentify images, file transfer management, clinical data management and assistance with high-performance computing machine learning/artificial intelligence.
The Research IT Department provides software tools, technologies and services to the research community for the development and implementation of biospecimen or data research studies, disease registries and research surveys. Offered are two fully supported vendor-based software products: REDCap and BayaTree RMS. Both applications provide a robust set of features and functionality that can be tailored to meet the unique research requirements of the investigators. Local, multi-site and international collaborations are supported with these tools. Our staff provides the knowledge and expertise for these technologies' design, development, testing, implementation, training and ongoing support.
Our team provides consultation services to the research community. We have a diverse set of knowledge, skills and expertise spanning a wide range of technologies and research domains. With this, we can provide insight, guidance, analysis, planning, design, methods and approach to meeting the technology needs of investigators and supporting the entire research study life cycle.
We provide statistical analysis tools for use throughout the research community. These tools are made available to anyone who has a need. These tools include SAS, SPSS, Stata, GraphPad and FCS Express. Our team manages the purchasing, licensing and implementation of these tools. Included with these tools is the EndNote application for bibliography management. We also provide procurement guidance and assistance to investigators when other analytic software packages are required.
Recent project highlights
nSight™ - Custom developed data discovery platform
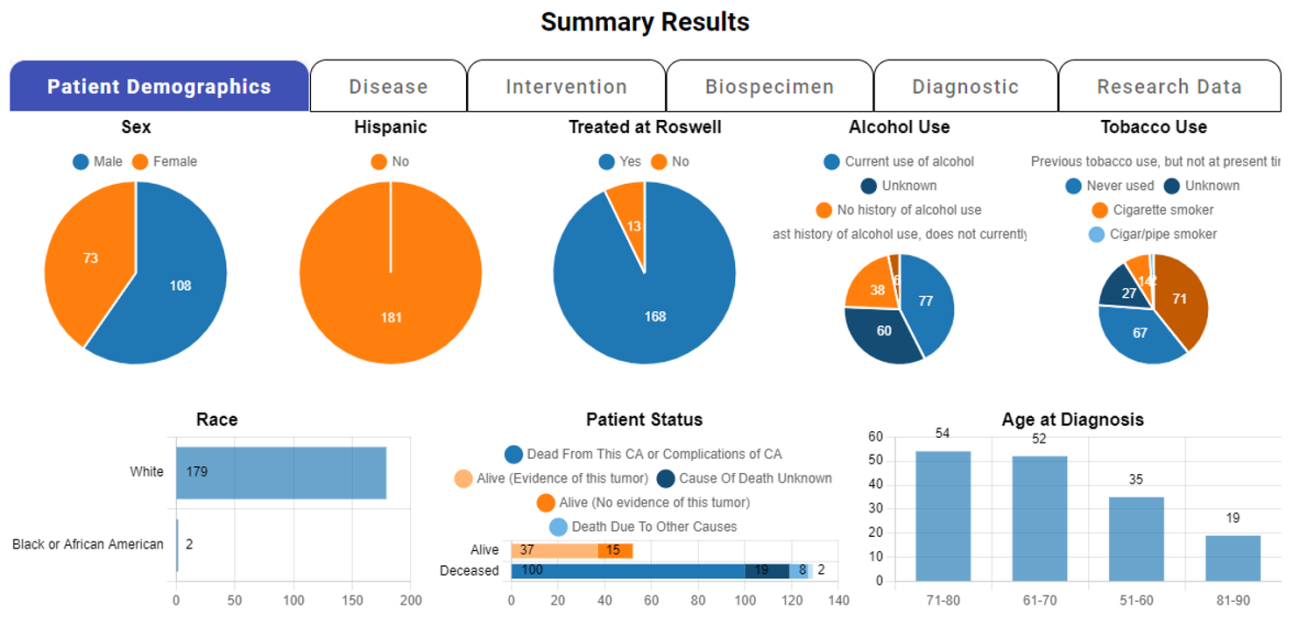
nSight™ is a data discovery platform provided by the Research Information Technology Department. This powerful system is made available for complimentary use by all Roswell Park staff who possess the requisite CITI training certifications. The core mission of the nSight initiative is to enable Roswell researchers with real-time access to vital clinical, research and regulatory data, thereby facilitating their research endeavors. With its diverse range of capabilities, the platform serves various purposes, including research feasibility assessment, grant proposal preparation, poster creation and clinical trial population design, among others.
Within this cutting-edge platform, users are empowered to conduct targeted searches for desired cases, utilizing a wide range of factors such as disease type, treatment modality and biospecimen characteristics.
Digital image research using artificial intelligence and machine learning
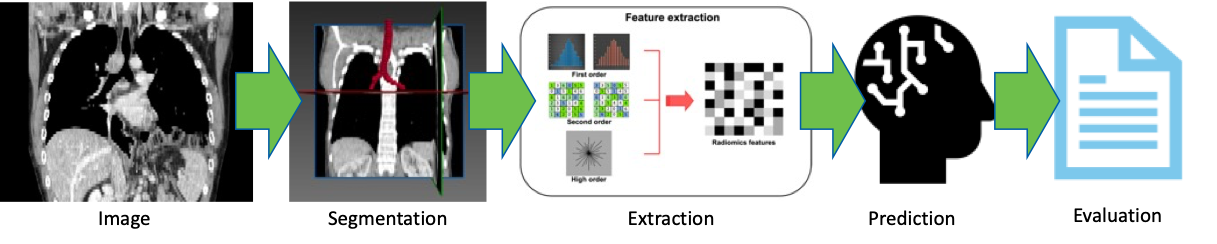
The Research IT Department is dedicated to harnessing the power of artificial intelligence (AI) and machine learning (ML) techniques to advance research in digital image analysis at Roswell Park. We believe that collaboration across multiple disciplines is essential for successful digital image research. The Research IT Department focuses on providing comprehensive support to researchers, enabling them to utilize AI and ML algorithms in the analysis of various types of digital images. These images include radiological images, pathology slides and histological images, which play a crucial role in diagnosis, prognosis and treatment planning for cancer patients.
The digital image research pipelines at Roswell Park typically encompass several key steps. Research IT facilitates image acquisition by collecting digital images from diverse sources. Next, we engage in annotation and segmentation, a process where specific regions of interest within the images are identified and marked, thus creating labeled data for training ML algorithms. Feature extraction is the subsequent step, involving the application of sophisticated algorithms to extract relevant characteristics from the images. These extracted features then serve as inputs for ML models. Finally, we carry out evaluation, which entails assessing the performance and accuracy of the ML models using appropriate metrics. This evaluation process ensures that the ML models are effective and contribute to the advancement of research in digital image analysis at Roswell Park.
The Research IT Department collaborates closely with researchers to ensure the seamless execution of these steps, with a focus on efficiency and effectiveness. Our aim is to drive innovative research outcomes and facilitate breakthroughs in the analysis of digital images. By leveraging AI and ML techniques, we strive to enhance the speed, accuracy, and precision of diagnoses, as well as the development of personalized treatment plans for cancer patients.
Location & hours
Roswell Park Comprehensive Cancer CenterResearch Information Technology Department
901 Washington St.
2nd Floor
Buffalo, New York 14263
Meet our team
Contact us
Wendy Raber
Department Administrator
Phone: 716-845-3936
Email: Wendy.Raber@RoswellPark.org